Engineering Complexity: Advanced 3D Bioprinting Strategies for Porous, Biomimetic Scaffolds in Regenerative Medicine
This article provides a comprehensive analysis of current 3D bioprinting techniques for fabricating porous, biomimetic scaffolds.
Engineering Complexity: Advanced 3D Bioprinting Strategies for Porous, Biomimetic Scaffolds in Regenerative Medicine
Abstract
This article provides a comprehensive analysis of current 3D bioprinting techniques for fabricating porous, biomimetic scaffolds. It explores the foundational principles of scaffold design, detailing key methodologies such as extrusion-based, inkjet, and light-assisted bioprinting. The content delves into practical applications in tissue engineering and drug screening, addresses common fabrication challenges and optimization strategies, and validates techniques through comparative analysis of structural, mechanical, and biological outcomes. Aimed at researchers and pharmaceutical developers, this review synthesizes cutting-edge advancements to guide the development of next-generation scaffolds for clinical translation.
The Blueprint of Life: Core Principles of Porosity and Biomimicry in Scaffold Design
Within the advancement of 3D bioprinting for porous biomimetic scaffolds, defining the ideal scaffold is paramount. It must transcend being a passive 3D structure and become a bioactive, instructive microenvironment that supports cell viability, proliferation, and specific function. This application note delineates the essential scaffold characteristics—architectural, mechanical, and biochemical—and provides detailed protocols for their quantitative assessment.
Essential Characteristics & Quantitative Data
A scaffold’s performance is governed by a hierarchy of physical and chemical properties. The following table summarizes key metrics and their target ranges for an ideal cell-supportive scaffold.
Table 1: Quantitative Characteristics of an Ideal Biomimetic Scaffold
| Characteristic | Parameter | Ideal Range/Target | Measurement Technique |
|---|---|---|---|
| Architectural | Porosity | > 90% | Micro-CT Analysis |
| Pore Size | 100 - 500 μm (cell-type dependent) | SEM Image Analysis | |
| Pore Interconnectivity | > 99% | Mercury Intrusion Porosimetry | |
| Mechanical | Compressive Modulus | 0.1 - 20 kPa (soft tissues) to 10 - 500 MPa (bone) | Uniaxial Compression Test |
| Degradation Rate | Match rate of neo-tissue formation (weeks-months) | Mass Loss in vitro | |
| Surface/Biochemical | Surface Roughness (Ra) | 0.5 - 5 μm | Atomic Force Microscopy |
| Bioactive Molecule Density | 0.1 - 1.0 μg/cm² | Fluorescent Tag Quantification | |
| Biological Performance | Cell Seeding Efficiency | > 85% | DNA Quantification Assay |
| Cell Viability (Day 7) | > 90% | Live/Dead Staining & Confocal | |
| Metabolic Activity (Day 7) | 150-300% of Day 1 Baseline | AlamarBlue/MTT Assay |
Experimental Protocols
Protocol 1: Assessment of Scaffold Porosity and Pore Architecture via Micro-CT Objective: To non-destructively quantify porosity, pore size distribution, and interconnectivity. Materials: µCT scanner (e.g., SkyScan 1272), scaffold sample (dry), image analysis software (CTAn, ImageJ). Procedure:
- Mount the dry scaffold securely on the sample stage.
- Set scan parameters: Voltage=40 kV, Current=250 µA, Pixel Size=5 µm, Rotation Step=0.4°, 180° rotation.
- Perform the scan and reconstruct cross-sectional slices using NRecon software.
- Import reconstructed slices into CTAn. Apply a uniform global threshold to binarize scaffold material from pores.
- Analysis:
- Porosity: Calculate as (Total Volume - Material Volume) / Total Volume.
- Pore Size Distribution: Execute the "Sphere Fitting" algorithm.
- Interconnectivity: Use the "Analyze Particles" function on a binarized inverse image; interconnectivity = (volume of connected pores / total pore volume) x 100%.
- Generate 3D models for visualization.
Protocol 2: Evaluation of Cell Viability, Distribution, and Metabolic Activity Objective: To assess the cytocompatibility and bioactivity of the scaffold over time. Materials: Seeded scaffold, PBS, Calcein-AM (4 µM) and Ethidium homodimer-1 (EthD-1, 2 µM) in PBS, AlamarBlue reagent, Confocal microscope, Plate reader. Procedure: Part A: Live/Dead Staining & Confocal Microscopy
- At designated time points (Days 1, 3, 7), aspirate culture medium.
- Rinse scaffold gently with warm PBS.
- Incubate with Live/Dead staining solution (Calcein-AM/ EthD-1) for 45 minutes at 37°C, protected from light.
- Rinse with PBS and image immediately using a confocal microscope (z-stack 50 µm deep). Calculate viability as: (Live cells / (Live+Dead cells)) x 100%. Part B: Metabolic Activity (AlamarBlue Assay)
- Prepare a 10% (v/v) solution of AlamarBlue reagent in fresh culture medium.
- After Live/Dead imaging, transfer scaffolds to a new plate, add the 10% reagent solution, and incubate for 3 hours at 37°C.
- Transfer 100 µL of the reacted supernatant to a 96-well plate.
- Measure fluorescence (Ex 560 nm / Em 590 nm) on a plate reader. Express data as percentage relative to Day 1 control scaffolds.
Protocol 3: Functional Assessment of Osteogenic Differentiation in a Bone Scaffold Objective: To quantify scaffold-induced osteogenic differentiation of human mesenchymal stem cells (hMSCs). Materials: hMSC-seeded scaffold, Osteogenic medium (OM: DMEM, 10% FBS, 10 mM β-glycerophosphate, 50 µM ascorbic acid-2-phosphate, 100 nM dexamethasone), ALP staining kit, Quantification buffer (pNPP), qPCR reagents for RUNX2, OSP. Procedure:
- Culture hMSC-seeded scaffolds in OM for 7, 14, and 21 days.
- Alkaline Phosphatase (ALP) Activity (Day 7/14):
- Lyse cells in 0.1% Triton X-100.
- Mix lysate with pNPP substrate, incubate 30 min, stop with 1N NaOH.
- Measure absorbance at 405 nm. Normalize to total DNA content.
- Gene Expression Analysis (Day 14/21):
- Extract total RNA, synthesize cDNA.
- Perform qPCR for osteogenic markers (RUNX2, OSP) and housekeeping gene (GAPDH). Calculate fold change using the 2^(-ΔΔCt) method.
Signaling Pathway: Integrin-Mediated Cell Adhesion on a Functionalized Scaffold
Title: RGD-Integrin Signaling to Cell Functions
Workflow: Bioprinting and Evaluating a Biomimetic Scaffold
Title: Bioprinted Scaffold Fabrication and Analysis Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Scaffold Biofabrication and Analysis
| Item | Function/Application | Example/Note |
|---|---|---|
| Gelatin Methacryloyl (GelMA) | Photo-crosslinkable bioink providing natural cell-adhesion motifs (RGD). | Tuneable mechanical properties via degree of substitution and concentration. |
| Poly(ethylene glycol) Diacrylate (PEGDA) | Synthetic, bio-inert hydrogel precursor for controlled mechanical and biochemical functionalization. | Serves as a blank slate for covalent attachment of peptides. |
| RGD Peptide Solution | Synthetic Arg-Gly-Asp peptides to functionalize synthetic scaffolds for integrin-mediated cell adhesion. | Commonly used at 0.5-2.0 mM concentration for coupling. |
| AlamarBlue Cell Viability Reagent | Resazurin-based, non-toxic assay for quantifying metabolic activity over time in the same sample. | Allows longitudinal tracking. |
| Calcein-AM / EthD-1 Live/Dead Kit | Two-color fluorescence assay for simultaneous determination of live (green) and dead (red) cells in 3D constructs. | Critical for confocal z-stack viability assessment. |
| Triton X-100 Solution (0.1%) | Non-ionic detergent for cell lysis to release intracellular enzymes (e.g., ALP) for quantitative biochemical assays. | |
| p-Nitrophenyl Phosphate (pNPP) | Colorimetric substrate for Alkaline Phosphatase (ALP), turning yellow upon cleavage, measured at 405 nm. | Standard for early osteogenic marker detection. |
| Osteogenic Induction Cocktail | Defined supplement (Dexamethasone, β-Glycerophosphate, Ascorbate) to direct hMSCs toward bone lineage. | Essential for functional differentiation assays. |
Application Notes
Within the field of 3D bioprinting for biomimetic scaffolds, porosity is not merely the presence of voids but a critical architectural determinant of biological function. The triad of interconnectivity, pore size, and diffusive transport dictates the ultimate success of a scaffold in supporting cell viability, tissue ingrowth, and functional maturation. This is paramount for applications in regenerative medicine, disease modeling, and drug screening.
- Interconnectivity: Refers to the degree to which pores are linked, forming a continuous network. It is the non-negotiable enabler for cell migration, vascularization, and uniform tissue formation. Scaffolds with high porosity but low interconnectivity act as isolated chambers, leading to necrotic cores.
- Pore Size: Governs specific cellular behaviors. While optimal sizes are cell-type and tissue-dependent, general ranges are established: <50µm for neovascularization, 50-150µm for osteogenesis, 100-350µm for bone ingrowth, and 200-500µm for chondrogenesis and hepatocyte function.
- Nutrient/Waste Diffusion: In the avascular phase post-implantation, survival depends on passive diffusion. The effective diffusion coefficient (Deff) within a porous scaffold is a function of its tortuosity (τ) and porosity (ε): Deff = (ε/τ) * D. This makes interconnectivity a direct regulator of metabolic waste clearance and nutrient supply, preventing central necrosis.
The design challenge in 3D bioprinting is to precisely engineer this porous architecture concurrently with the deposition of bioinks. Techniques like sacrificial bioprinting, cryogenic printing, and the tuning of printing parameters (pressure, speed, infill pattern) are leveraged to achieve target porosity profiles.
Table 1: Optimal Pore Size Ranges for Tissue-Specific Scaffolds
| Tissue Target | Optimal Pore Size Range (µm) | Primary Cellular/Physiological Rationale |
|---|---|---|
| Bone Regeneration | 300 - 500 | Facilitates osteoconduction, vascular invasion, and bone matrix deposition. |
| Cartilage Tissue | 200 - 350 | Supports chondrocyte migration and ECM production, while maintaining structural integrity. |
| Skin Regeneration | 80 - 150 | Promotes fibroblast infiltration, keratinocyte migration, and rapid neovascularization. |
| Neural Tissue | 50 - 120 | Guides neurite extension and supports glial cell integration. |
| Hepatocyte Culture | 200 - 500 | Enhances oxygenation and mass transfer for high-density, metabolically active cells. |
| Vascular Ingrowth | > 50 (interconnected) | Minimum size required for endothelial cell sprouting and capillary formation. |
Table 2: Impact of Porosity & Interconnectivity on Diffusion Efficiency
| Scaffold Material | Total Porosity (ε) % | Interconnectivity (%) | Measured D_eff / D (Glucose) | Key Outcome (In Vitro/In Vivo) |
|---|---|---|---|---|
| 3D Printed PCL (Honeycomb) | 70 | ~100 | 0.68 | Uniform cell distribution; no central necrosis after 14 days. |
| Salt-Leached PLGA | 85 | 75 | 0.45 | Viable periphery, necrotic core (>500µm depth) observed. |
| Gelatin Methacryloyl (GelMA) | 90 (Cryogel) | >95 | 0.82 | Superior cell viability (>95%) throughout 1.5mm thick construct. |
| Silk Fibroin (Freeze-dried) | 92 | 60 | 0.32 | Limited cell infiltration; viability restricted to <200µm surface layer. |
Experimental Protocols
Protocol 1: Quantifying Scaffold Interconnectivity via Micro-CT Analysis
Objective: To non-destructively calculate the percentage of interconnected porosity within a 3D-bioprinted scaffold. Materials: Micro-CT scanner (e.g., SkyScan 1272), 3D-bioprinted scaffold sample (dry), image analysis software (CTAn, Fiji/ImageJ), computer. Procedure:
- Sample Mounting: Secure the dry scaffold on the sample holder using low-density foam to prevent movement.
- Scanning Parameters: Set isotropic voxel size to 1/3 of the target minimum pore diameter. Use a rotation step of 0.4° over 180°. Apply a 0.5mm aluminum filter if using a polymer scaffold to reduce beam hardening.
- Image Reconstruction: Use the scanner's proprietary software (NRecon for SkyScan) to reconstruct cross-sectional images. Apply consistent beam hardening and ring artifact correction.
- Binarization (CTAn):
- Load the image stack. Define a global threshold using the Otsu method to separate solid material from pore space.
- Apply a despeckle function to remove noise.
- 3D Analysis:
- Select the region of interest (ROI) encompassing the entire scaffold.
- Execute "Analysis -> 3D Analysis." The key output is "Open Porosity %," which represents the interconnected pore volume as a percentage of total scaffold volume. The "Closed Porosity %" represents isolated pores.
- Visualization: Use CTVox or Fiji's 3D Viewer to generate 3D models of the interconnected pore network.
Protocol 2: Evaluating Nutrient Diffusion and Cell Viability in Thick Scaffolds
Objective: To correlate scaffold porosity parameters with gradients in cell viability driven by diffusion limits. Materials: 3D-bioprinted cell-laden scaffold (e.g., with mesenchymal stem cells), live/dead viability/cytotoxicity kit (Calcein AM/EthD-1), confocal microscope, image analysis software (e.g., Imaris, Fiji). Procedure:
- Scaffold Culture: Culture a thick (>2mm) cell-laden scaffold in standard medium for 3-7 days under static conditions to establish diffusion-driven gradients.
- Staining:
- Prepare staining solution per manufacturer's protocol (e.g., 2µM Calcein AM, 4µM Ethidium homodimer-1 in PBS).
- Rinse scaffold with PBS. Incubate in staining solution for 45-60 minutes at 37°C, protected from light.
- Imaging:
- Rinse and image the scaffold immediately. Using a confocal microscope, acquire Z-stacks from the top surface to the center of the scaffold. Use 10x or 20x objective.
- Set excitation/emission for Calcein (494/517 nm, green) and EthD-1 (528/617 nm, red).
- Quantitative Analysis (Fiji):
- Split channels. For each Z-slice (representing a depth), threshold the green (live) and red (dead) channels separately.
- Measure the area fraction of live and dead signal for each slice.
- Plot % Live Cells vs. Depth (µm).
- Correlation: Compare the depth at which viability drops below 70% with the scaffold's measured interconnectivity and average pore size from Protocol 1. High-interconnectivity scaffolds will maintain viability to greater depths.
Diagrams
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Porosity & Diffusion Studies
| Item | Function in Research | Key Consideration |
|---|---|---|
| Sacrificial Bioink (e.g., Pluronic F-127, Gelatin) | Printed as a temporary lattice to create interconnected channels, which are later liquefied and removed. | Must be cytocompatible during printing and dissolve under mild conditions (e.g., cooling, enzymatic digestion). |
| Micro-Computed Tomography (Micro-CT) System | Provides non-destructive 3D quantification of total porosity, pore size distribution, and interconnectivity. | Resolution must be significantly higher than the feature size of interest (e.g., <5µm voxel for 50µm pores). |
| Live/Dead Viability/Cytotoxicity Kit | Dual-fluorescence stain (Calcein AM for live, EthD-1 for dead cells) to map viability gradients within scaffolds. | Requires penetration through the scaffold matrix; incubation times may need optimization for dense constructs. |
| Mathematical Diffusion Models (e.g., Fick's Law, COMSOL) | Used to predict nutrient/waste concentration gradients and D_eff based on scaffold architecture pre-experiment. | Input parameters (ε, τ) must be derived from accurate structural data (e.g., Micro-CT). |
| Tunable Hydrogel (e.g., GelMA, Alginate) | Base bioink material whose crosslinking density can be modulated to intrinsically alter matrix porosity and diffusivity. | Polymer concentration, degree of methacrylation, and UV crosslinking time are critical control parameters. |
| Perfusion Bioreactor System | Applied post-printing to overcome diffusion limits, allowing culture of large, dense constructs. Validates the importance of designed porosity. | Flow rates must be optimized to provide shear stress conducive to the target cell type without causing detachment. |
Within the broader thesis on 3D bioprinting techniques for porous biomimetic scaffolds, replicating native tissue ECM architecture is paramount. The ECM is not a passive scaffold but a dynamic, instructive niche that regulates cellular adhesion, migration, proliferation, differentiation, and mechanotransduction. Biomimicry of the ECM involves recapitulating its biochemical composition, hierarchical architecture (from nano- to microscale), mechanical properties, and spatiotemporal signaling. This document provides application notes and protocols for analyzing native ECM and fabricating biomimetic scaffolds via advanced 3D bioprinting.
Quantitative Analysis of Native ECM Architecture
Understanding native ECM parameters is the first critical step. Below are summarized metrics for key tissues, guiding scaffold design targets.
Table 1: Architectural and Mechanical Properties of Native Tissue ECM
| Tissue Type | Average Fiber Diameter (nm) | Pore Size Range (μm) | Porosity (%) | Elastic Modulus (kPa) | Predominant ECM Components |
|---|---|---|---|---|---|
| Skin (Dermis) | 50 - 150 | 50 - 200 | 75 - 90 | 2 - 80 | Collagen I/III, Elastin, HA, Fibronectin |
| Bone (Trabecular) | 500 - 2000 (fibril bundle) | 200 - 600 | 50 - 90 | 10,000 - 20,000 | Collagen I, Hydroxyapatite |
| Articular Cartilage | 20 - 80 | 30 - 100 | 60 - 85 | 500 - 1000 | Collagen II, Aggrecan, HA |
| Liver | 40 - 100 | 10 - 50 | 20 - 40 | 0.5 - 1.5 | Collagen I/III/IV, Laminin, Fibronectin |
| Cardiac Muscle | 80 - 200 | 50 - 150 | 70 - 80 | 10 - 50 | Collagen I/IV, Laminin, Fibronectin |
| Peripheral Nerve | 50 - 100 (basal lamina) | 5 - 20 (conduit) | 60 - 80 | 0.5 - 1.0 | Collagen IV, Laminin, Fibronectin |
Core Experimental Protocols
Protocol 3.1: Decellularized ECM (dECM) Hydrogel Preparation for Bioink Formulation
Objective: To create a bioactive bioink derived from tissue-specific ECM. Materials: See Scientist's Toolkit, Section 5.0. Procedure:
- Tissue Acquisition & Processing: Mince 1g of porcine or human source tissue (e.g., skin, heart) into <1 mm³ pieces. Rinse in PBS with 1% Antibiotic-Antimycotic.
- Decellularization: Treat tissue with:
- 0.1% SDS for 48h (agitation at 4°C) to lyse cells and solubilize cytoplasmic components.
- Rinse with PBS until no SDS is detectable (conductivity measurement).
- Treat with 1% Triton X-100 for 24h (agitation, RT) to remove nuclear remnants.
- Rinse with nuclease-free water (72h, with changes) to remove detergents.
- Lyophilization & Milling: Freeze at -80°C, lyophilize for 48h. Pulverize into a fine powder using a cryomill.
- Digestion & pH Neutralization: Digest dECM powder in 0.5M acetic acid with 1 mg/mL pepsin (w/v) for 48-72h at 4°C (constant stirring). Maintain a 1:100 (w/v) ratio. Neutralize to pH 7.4 using 1M NaOH and dilute with 10x PBS to achieve isotonicity.
- Sterilization & Storage: Filter sterilize (0.22 μm) under vacuum. Store at 4°C for up to 1 week. For long-term storage, aliquot and freeze at -80°C.
Protocol 3.2: Multi-Material 3D Bioprinting of a Zonal Cartilage-Mimetic Scaffold
Objective: To fabricate a scaffold mimicking the depth-dependent collagen architecture of articular cartilage. Materials: See Scientist's Toolkit, Section 5.0. Procedure:
- Bioink Preparation:
- Superficial Zone Ink: 8% (w/v) Methacrylated Silk Fibroin (SilMA), 2% (w/v) Methacrylated Hyaluronic Acid (HAMA), 0.1% LAP photoinitiator. Mix with primary chondrocytes at 20x10⁶ cells/mL.
- Deep Zone Ink: 4% (w/v) SilMA, 4% (w/v) Methacrylated Chondroitin Sulfate (CSMA), 0.1% LAP. Mix with chondrocytes at 10x10⁶ cells/mL.
- Printing Parameters (Extrusion-based, dual-head):
- Nozzle Diameter: 250 μm.
- Pressure: 20-25 kPa (Superficial), 15-20 kPa (Deep).
- Print Speed: 8 mm/s.
- Print Bed Temp: 15°C.
- Layer-by-Layer Fabrication:
- Program a 0/90° filament pattern for each layer.
- Print 10 layers of Superficial Zone Ink (final height ~500 μm).
- Without interrupting the print, switch to Deep Zone Ink.
- Print subsequent 40 layers (final scaffold height ~2.5 mm).
- Crosslinking: Immediately after printing, expose the entire construct to 405 nm light at 15 mW/cm² for 60 seconds per side. Incubate in chondrogenic medium (with TGF-β3) at 37°C, 5% CO₂.
Key Signaling Pathways in ECM-Biomimetic Scaffold Interaction
Diagram 1: Key Mechanotransduction Pathways from Biomimetic ECM
Diagram 2: Workflow for Biomimetic ECM Scaffold Development
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for ECM Biomimicry and 3D Bioprinting
| Item Name / Catalog Example | Category | Primary Function in Research |
|---|---|---|
| Methacrylated Gelatin (GelMA) | Bioink Polymer | Provides RGD motifs for cell adhesion and tunable photocrosslinkability for soft tissue mimics. |
| Decellularized ECM (dECM) Powder | Bioink Additive | Preserves tissue-specific biochemical cues (collagens, GAGs, growth factors). |
| Lithium Phenyl-2,4,6-Trimethylbenzoylphosphinate (LAP) | Photoinitiator | Enables rapid, cytocompatible visible light (405 nm) crosslinking of methacrylated bioinks. |
| Sodium Dodecyl Sulfate (SDS) | Detergent | Primary agent for decellularization; lyses cells and solubilizes cytoplasmic components. |
| Triton X-100 | Detergent | Non-ionic surfactant used to remove nuclear remnants and lipids after SDS treatment. |
| Recombinant Human TGF-β3 | Growth Factor | Induces chondrogenic differentiation in mesenchymal stem cells for cartilage scaffolds. |
| Integrin α5β1 Inhibitor (ATN-161) | Signaling Modulator | Validates the specific role of fibronectin-integrin interactions in cell-scaffold responses. |
| AlamarBlue / CellTiter-Glo | Viability Assay | Quantifies metabolic activity and proliferation of cells within 3D printed constructs. |
| Human Dermal Fibroblasts (HDFs) or Mesenchymal Stem Cells (MSCs) | Cell Source | Standardized primary cells for evaluating scaffold biocompatibility and bioactivity. |
| Advanced Rheometer (e.g., TA Instruments) | Equipment | Characterizes bioink viscoelastic properties (storage/loss modulus) to optimize printability. |
Application Notes and Protocols
Thesis Context: Within the research on 3D bioprinting techniques for porous biomimetic scaffolds, the selection and formulation of the biomaterial ink (bioink) is critical. These scaffolds must mimic the extracellular matrix (ECM), provide structural integrity, support cell viability, and guide tissue regeneration. This document details the application and processing protocols for key biomaterial classes.
Hydrogel-Based Bioinks: Crosslinking Protocols
Hydrogels form the foundational aqueous environment for cell encapsulation. Their crosslinking method dictates printability, mechanical properties, and gelation kinetics.
Protocol 1.1: Dual-Crosslinking of Alginate-Gelatin (Alg-Gel) Composite Bioink
Objective: To create a shape-fidelity stable, cell-laden construct using ionic and thermal crosslinking. Materials: See "The Scientist's Toolkit," Table 2. Workflow:
- Bioink Preparation: Sterilize sodium alginate (4% w/v) and gelatin (8% w/v) in DPBS separately at 60°C. Mix in a 1:1 ratio under sterile conditions. Cool to 37°C and mix with cell suspension to a final density of 1-5 x 10^6 cells/mL.
- Printing & Primary Crosslink: Bioprint the Alg-Gel/cell mixture into a pre-cooled (4°C) printing bed. The rapid thermal gelation of gelatin provides initial shape fidelity.
- Secondary Crosslink: Immerse the printed construct in a sterile 100 mM CaCl₂ solution for 5-10 minutes. Ca²⁺ ions ionically crosslink the alginate chains, providing long-term stability.
- Culture: Rinse with culture medium and transfer to an incubator (37°C, 5% CO₂). Gelatin gradually solubilizes, increasing scaffold porosity and leaving behind a stable alginate network.
Diagram: Dual-Crosslinking Workflow for Alg-Gel Bioink
Table 1: Crosslinking Methods for Common Hydrogel Bioinks
| Hydrogel | Crosslinking Mechanism | Key Agent/Stimulus | Gelation Time | Key Property |
|---|---|---|---|---|
| Alginate | Ionic | Ca²⁺, Ba²⁺ ions | Seconds-Minutes | Rapid, reversible |
| GelMA | Photo-chemical | UV/VIS light (LAP photoinitiator) | < 60 seconds | Spatiotemporal control |
| Fibrin | Enzymatic | Thrombin + Ca²⁺ | Seconds | Natural cell adhesion |
| PEGDA | Photo-chemical | UV light | Seconds | Highly tunable mechanics |
| Collagen I | Thermal/pH | Temperature shift to 37°C, pH 7.4 | Minutes | Native ECM composition |
Ceramic Bioinks: Printing and Sintering for Bone Scaffolds
Ceramics like hydroxyapatite (HA) and tricalcium phosphate (TCP) are essential for osteoconductive bone scaffolds but require dispersion in a carrier hydrogel for printability.
Protocol 2.1: Direct Ink Writing (DIW) of α-TCP/Pluronic F127 Composite Paste Objective: To fabricate a macro-porous ceramic green body for subsequent sintering into a pure bioceramic scaffold. Materials: α-TCP powder (≤ 50 µm), Pluronic F127, deionized water, 3D bioprinter with pneumatic syringe extruder, high-temperature furnace. Workflow:
- Paste Synthesis: Create a 30% w/v Pluronic F127 solution in cold water. Gradually blend in α-TCP powder to a final ceramic loading of 40-50% w/w. Homogenize under vacuum to remove air bubbles.
- Printing: Load paste into a syringe. Print at 4-10°C (to keep Pluronic viscous) through a tapered nozzle (400-610 µm). Apply constant pressure (25-40 psi) for steady extrusion.
- Post-Processing: Air-dry the printed "green body" for 24h. Sinter in a furnace with a controlled ramp (1-5°C/min to 1100-1300°C, hold for 2h, cool slowly). Pluronic burns off, leaving a densified, micro-porous α-TCP scaffold.
Table 2: Sintering Parameters for Ceramic Bioinks
| Ceramic Type | Sintering Temp. Range | Hold Time | Final Porosity | Compressive Strength |
|---|---|---|---|---|
| β-Tricalcium Phosphate (β-TCP) | 1100 - 1150°C | 2-3 hours | 40-60% | 10-20 MPa |
| Hydroxyapatite (HA) | 1200 - 1300°C | 2-4 hours | 30-50% | 20-50 MPa |
| Bioactive Glass (4555) | 600 - 700°C | 1 hour | 50-70% | 5-15 MPa |
Composite Bioink Protocol: Reinforced GelMA with Nanocellulose
Protocol 3.1: Formulating a Nanocomposite Bioink for Meniscus Tissue Engineering Objective: To enhance the mechanical resilience of a soft, cell-laden hydrogel for load-bearing soft tissue applications. Materials: GelMA (5-10% w/v), Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) photoinitiator (0.25% w/v), TEMPO-oxidized cellulose nanofibrils (CNF, 0.5-1.5% w/v), chondrocytes. Workflow:
- CNF Dispersion: Sonicate CNF suspension in PBS (1% w/v) for even dispersion.
- Bioink Formulation: Dissolve GelMA and LAP in warm PBS (40°C). Cool to room temp, then mix with the CNF dispersion and cell suspension. Final concentrations: 7.5% GelMA, 0.25% LAP, 1% CNF, 5x10^6 cells/mL.
- Printing & Crosslinking: Extrude bioink into a lattice structure. Immediately expose to 405 nm visible light (5-10 mW/cm²) for 60-90 seconds per layer for full photocrosslinking.
- Mechanical Assessment: Perform rheology (storage modulus G') and cyclical compression testing to quantify reinforcement.
Diagram: CNF-Reinforced GelMA Composite Mechanism
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagents for Bioink Development and Characterization
| Reagent/Material | Function & Rationale | Example Vendor/Product |
|---|---|---|
| Gelatin Methacryloyl (GelMA) | Photo-crosslinkable hydrogel providing natural RGD motifs for cell adhesion. | Advanced BioMatrix, ESI-BIO |
| Lithium Phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Cytocompatible, visible-light photoinitiator for rapid crosslinking of GelMA/PEGDA. | Sigma-Aldrich, TCI Chemicals |
| Alginic Acid (Sodium Salt) | Polysaccharide for ionic crosslinking; provides shear-thinning behavior. | NovaMatrix, Sigma-Aldrich |
| β-Tricalcium Phosphate (β-TCP) Powder | Osteoconductive ceramic for bone scaffold bioinks. | Sigma-Aldrich, Berkeley Advanced Biomaterials |
| TEMPO-Oxidized Nanocellulose | Nanofibrillar additive to enhance bioink viscosity, stiffness, and shape fidelity. | CelluForce, University of Maine Process Center |
| Pluronic F127 | Thermoreversible sacrificial polymer for ceramic paste printing. | Sigma-Aldrich, BASF |
| Human Mesenchymal Stem Cells (hMSCs) | Multipotent primary cells for evaluating osteogenic/chondrogenic differentiation in scaffolds. | Lonza, ATCC |
| Live/Dead Viability/Cytotoxicity Kit | Fluorescent assay (Calcein AM/EthD-1) to quantify cell viability post-printing. | Thermo Fisher Scientific |
The success of 3D bioprinted porous biomimetic scaffolds in tissue engineering hinges on the recapitulation of native biological imperatives. This involves the precise spatial and temporal incorporation of three core components: (1) living cells as building blocks, (2) growth factors (GFs) as biochemical messengers, and (3) physical/chemical signaling cues from the scaffold matrix. The convergence of these elements within a porous 3D architecture directs cell fate—including adhesion, proliferation, migration, and differentiation—toward functional tissue formation. For researchers and drug development professionals, mastering this integration is critical for developing advanced in vitro disease models, high-throughput drug screening platforms, and clinically viable implants.
Key Application Notes:
- Spatial Patterning: Multi-material bioprinting allows the codeposition of different cell types and bioinks laden with specific GFs into distinct zones of a scaffold, mimicking tissue interfaces (e.g., osteochondral tissue).
- Temporal Control: GF release kinetics can be engineered via encapsulation in microparticles (e.g., gelatin, PLGA) or by leveraging enzyme-sensitive or shear-thinning bioinks to provide staged signaling crucial for complex morphogenesis.
- Mechanotransduction: Scaffold porosity, stiffness (elastic modulus), and microtopography are physical cues that interact with integrin-mediated signaling pathways, influencing stem cell lineage commitment. A pore size of 100-300 µm is often optimal for cell infiltration and vascularization.
- Drug Screening: Bioprinted tumor models incorporating patient-derived cells, relevant ECM components, and gradient-forming GF cues provide more predictive platforms for oncology drug efficacy and toxicity testing compared to 2D cultures.
Table 1: Common Growth Factors & Their Roles in 3D Bioprinted Scaffolds
| Growth Factor | Abbreviation | Typical Concentration Range in Bioinks | Primary Cellular Target | Key Function in 3D Scaffolds |
|---|---|---|---|---|
| Vascular Endothelial Growth Factor | VEGF | 10-100 ng/mL | Endothelial Cells, Progenitors | Promotes angiogenesis; critical for vascularization of thick constructs. |
| Bone Morphogenetic Protein-2 | BMP-2 | 50-200 ng/mL | Mesenchymal Stem Cells (MSCs), Osteoprogenitors | Induces osteogenic differentiation; enhances bone regeneration. |
| Basic Fibroblast Growth Factor | bFGF/FGF-2 | 5-50 ng/mL | MSCs, Chondrocytes, Fibroblasts | Promotes cell proliferation, chondrogenesis, and maintains stemness. |
| Transforming Growth Factor-beta 3 | TGF-β3 | 10-50 ng/mL | MSCs, Chondrocytes | Potent inducer of chondrogenic differentiation; stimulates ECM production. |
| Nerve Growth Factor | NGF | 50-100 ng/mL | Neurons, PC12 Cells | Supports neuronal outgrowth, guidance, and survival in neural scaffolds. |
Table 2: Impact of Scaffold Physical Cues on Cell Behavior
| Scaffold Parameter | Typical Target Range | Key Signaling Pathways Influenced | Cellular Outcome |
|---|---|---|---|
| Average Pore Size | 100-300 µm (bone), 150-500 µm (soft tissue) | Integrin/FAK, YAP/TAZ | Cell infiltration, nutrient diffusion, neo-tissue formation. |
| Elastic Modulus (Stiffness) | ~0.1-1 kPa (brain), ~8-15 kPa (muscle), ~25-40 kPa (bone) | Rho/ROCK, YAP/TAZ, MKL/SRF | Stem cell lineage specification (soft->neuro/adirogenic, stiff->osteogenic). |
| Fiber Diameter (Nanofibrous) | 50-500 nm | Integrin clustering, Wnt/β-catenin | Enhanced cell adhesion, spreading, and differentiation. |
Experimental Protocols
Protocol 1: Bioprinting a VEGF-Gradient Scaffold for Angiogenesis Studies Aim: To fabricate a gelatin methacryloyl (GelMA)-based scaffold with a spatially defined VEGF gradient to direct endothelial cell network formation.
Materials:
- Cells: Human Umbilical Vein Endothelial Cells (HUVECs).
- Bioink A: 7% (w/v) GelMA, 0.25% photoinitiator (LAP), HUVECs (5x10^6 cells/mL) in cell culture medium.
- Bioink B: 7% (w/v) GelMA, 0.25% LAP, HUVECs (5x10^6 cells/mL), VEGF (50 ng/mL) in medium.
- Bioprinter: Extrusion-based with dual-printhead and UV crosslinking module.
- Software: Slicer capable of generating gradient infill patterns.
Method:
- Preparation: Sterilize printer stage and printheads. Maintain cells and bioinks on ice.
- Bioink Loading: Load Bioink A into printhead 1 and Bioink B into printhead 2.
- Gradient Design: Using the slicer software, design a rectangular construct (10x10x2 mm). Set the infill pattern to a linear gradient from 100% Bioink A (left) to 100% Bioink B (right).
- Printing Parameters: Set nozzle diameter (27G), pressure (20-30 kPa), printing speed (5 mm/s), and layer height (150 µm). Stage temperature: 15°C.
- Bioprinting: Print the construct layer-by-layer. After each layer, apply UV light (365 nm, 5 mW/cm²) for 30 seconds for partial crosslinking.
- Final Crosslinking: Post-print, irradiate the entire construct with UV light (365 nm, 5 mW/cm²) for 2 minutes.
- Culture: Transfer to endothelial cell growth medium (without VEGF) and culture for up to 14 days. Analyze network formation (total tube length, junctions) via fluorescence microscopy on days 3, 7, and 14.
Protocol 2: Assessing Osteogenic Differentiation in BMP-2 Loaded, Bioprinted Scaffolds Aim: To evaluate the osteo-inductive capacity of a BMP-2-releasing, alginate/hydroxyapatite bioink on encapsulated MSCs.
Materials:
- Cells: Human Bone Marrow-derived MSCs (passage 3-5).
- Bioink: 3% (w/v) alginate, 2% (w/v) nano-hydroxyapatite, 5x10^6 MSCs/mL, BMP-2 (100 ng/mL).
- Crosslinker: 100 mM CaCl₂ solution.
- Control: Bioink without BMP-2.
Method:
- Scaffold Fabrication: Extrude bioink into a grid structure (15x15x3 mm) directly into a CaCl₂ bath for 10 minutes for ionic crosslinking. Wash with PBS.
- Culture: Maintain scaffolds in basic osteogenic medium (DMEM, 10% FBS, 1% Pen/Strep) without dexamethasone or β-glycerophosphate to isolate BMP-2 effect. Change media every 3 days.
- Analysis (Day 21):
- Gene Expression (qRT-PCR): Lyse cells, extract RNA, and analyze markers: Runx2 (early), Osteocalcin (OCN) (late). Normalize to GAPDH.
- Protein Detection (Immunofluorescence): Fix scaffolds, permeabilize, stain for OCN (primary anti-OCN, secondary Alexa Fluor 488), and DAPI for nuclei. Image via confocal microscopy.
- Biochemical Assay (Quantitative): Perform Alkaline Phosphatase (ALP) activity assay on cell lysates using pNPP substrate. Normalize to total protein content (BCA assay).
Diagrams
Diagram 1: Key Signaling Pathways in a 3D Bioprinted Scaffold
Diagram 2: Workflow for a Functionalized Bioink Experiment
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Incorporating Biological Imperatives
| Item | Function in 3D Bioprinting Research | Example(s) |
|---|---|---|
| Photo-crosslinkable Hydrogels | Provide a biocompatible, printable matrix that can be stabilized via light, enabling high-fidelity 3D structures. | Gelatin Methacryloyl (GelMA), Poly(ethylene glycol) Diacrylate (PEGDA). |
| Integrin-Binding Peptides | Graft onto inert polymers to provide essential cell adhesion signaling (e.g., via RGD sequence). | RGD (Arg-Gly-Asp) peptide, IKVAV peptide for neural cells. |
| Growth Factor Carriers | Protect GFs from denaturation and control their release kinetics from the scaffold over time. | Gelatin microparticles, Heparin-based coacervates, PLGA nanoparticles. |
| Mechano-sensitive Reporters | Tools to visualize and quantify cellular response to scaffold physical properties. | FRET-based tension biosensors (e.g., Vinculin-FRET), YAP/TAZ localization antibodies. |
| Viability/Cytotoxicity Assay Kits | Standardized methods to assess cell health post-printing in 3D. | Live/Dead (Calcein AM/EthD-1) assay, AlamarBlue/Resazurin metabolic assay. |
| Decellularized ECM (dECM) Bioinks | Provide a complex, tissue-specific cocktail of native structural proteins and signaling molecules. | Cardiac dECM, Liver dECM, Adipose dECM powders. |
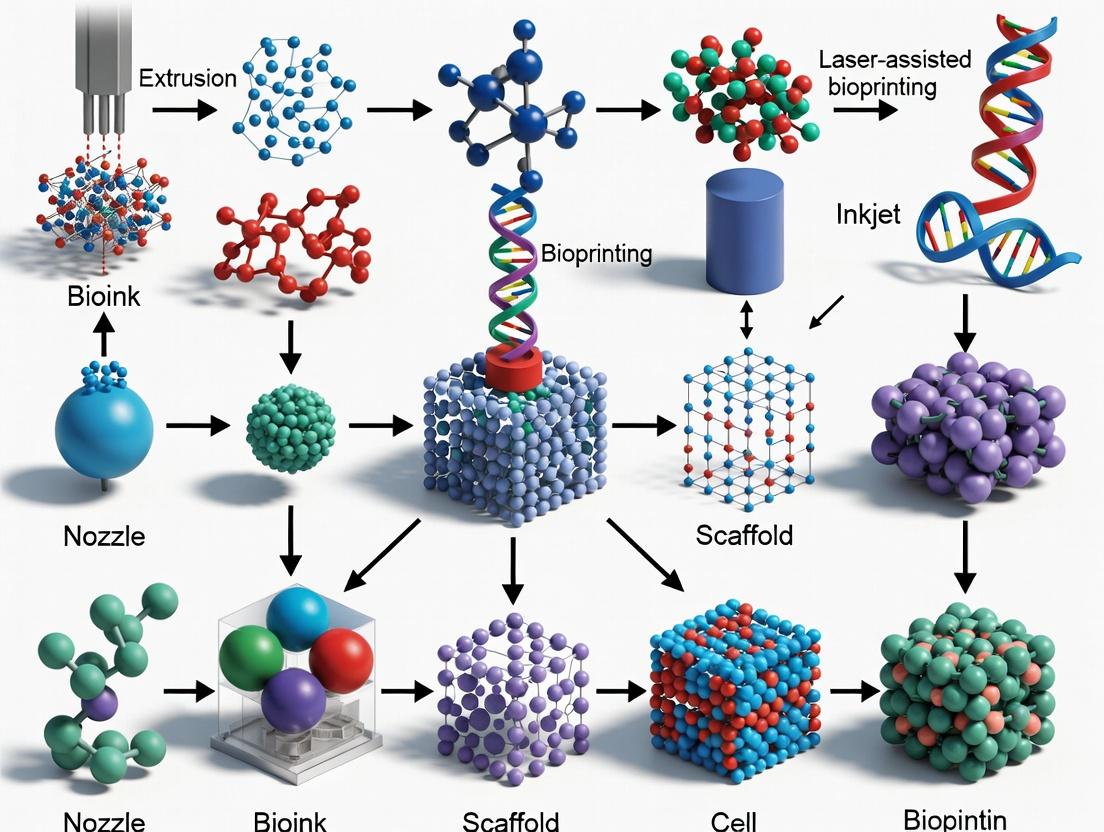
From Digital Model to Living Construct: A Guide to Primary 3D Bioprinting Techniques
Within the broader thesis on 3D bioprinting techniques for fabricating porous biomimetic scaffolds, extrusion-based bioprinting (EBB) stands out as the predominant and versatile workhorse. Its significance lies in its unique capacity to handle high-viscosity, cell-laden bioinks, enabling the fabrication of high-cell-density constructs essential for tissue engineering and drug development. This application note details protocols and data central to leveraging EBB for creating biomimetic, cell-rich porous structures.
Key Advantages and Quantitative Performance Data
The following table summarizes the core capabilities of EBB that make it ideal for high-cell-density applications.
Table 1: Performance Metrics of Extrusion-Based Bioprinting for High-Cell-Density Constructs
| Parameter | Typical Range/Value | Significance for High-Cell-Density Constructs |
|---|---|---|
| Bioink Viscosity | 30 mPa·s to > 6 x 10⁷ mPa·s | Enables incorporation of high cell densities (10⁶ – 10⁸ cells/mL) without rapid sedimentation. |
| Cell Viability (Post-Printing) | 40% – 95% (Protocol Dependent) | Direct function of shear stress management; target is >80% for functional constructs. |
| Printing Resolution | 50 µm – 1 mm | Suitable for creating porous networks (100-500 µm pores) that mimic native tissue vasculature. |
| Printing Speed | 1 – 50 mm/s | Balances throughput with shear stress exposure time. |
| Maximum Cell Density | Up to 1 x 10⁸ cells/mL | Facilitates fabrication of tissue-like cellularity, crucial for organotypic models and tissue repair. |
| Common Gelation Method | Thermal, Ionic, Photocrosslinking | Provides structural integrity to the soft, cell-dense filament post-deposition. |
Detailed Experimental Protocols
Protocol 1: Bioink Preparation & Rheological Characterization for High Cell Density
Aim: To formulate and characterize a shear-thinning bioink capable of supporting high cell density.
Materials:
- Primary cells or cell line (e.g., NIH/3T3, hMSCs)
- Base hydrogel (e.g., Alginate, GelMA, Collagen, Fibrin)
- Crosslinker (e.g., CaCl₂ for alginate, photoinitiator for GelMA)
- Cell culture medium (appropriate for cell type)
- Sterile centrifuge tubes, pipettes, biosafety cabinet
Method:
- Cell Harvesting: Culture cells to ~80% confluence. Trypsinize, quench, and centrifuge. Count cells using a hemocytometer.
- Bioink Mixing: Prepare sterile hydrogel precursor solution. Gently resuspend the cell pellet in the precursor solution to achieve the target final density (e.g., 5 x 10⁶ to 5 x 10⁷ cells/mL). Mix by slow pipetting to minimize air bubbles and shear stress. Keep on ice for temperature-sensitive materials (e.g., collagen).
- Rheology:
- Load bioink onto a parallel-plate rheometer.
- Perform a shear rate sweep (e.g., 0.1 to 100 s⁻¹) at printing temperature to confirm shear-thinning behavior.
- Perform an amplitude sweep to determine the linear viscoelastic region and gelation time if applicable.
- Key Metric: Ensure viscosity at high shear (∼10-100 s⁻¹, simulating nozzle flow) is low enough for extrusion, and at low shear (∼0.1 s⁻¹, post-deposition) is high enough to hold shape.
Protocol 2: Optimized Extrusion Bioprinting of High-Density Constructs
Aim: To print a porous, high-cell-density scaffold with maintained cell viability.
Materials:
- Prepared cell-laden bioink
- Extrusion bioprinter (pneumatic or piston-driven)
- Sterile printing cartridge and nozzles (diameter: 22G-27G)
- Sterile, crosslinking-compatible substrate (e.g., Petri dish, transwell)
- Crosslinking apparatus (e.g., UV lamp for GelMA, CaCl₂ mist for alginate)
Method:
- Printer Setup: Sterilize all fluid-path components (nozzle, cartridge) with 70% ethanol and UV light. Load the bioink into the cartridge, avoiding bubbles. Mount cartridge and nozzle.
- Print Parameter Calibration:
- Set printing temperature (e.g., 4°C for collagen, 20-37°C for others).
- Calibrate pressure (pneumatic) or plunger speed (piston) using a dummy bioink of similar viscosity to achieve a consistent filament flow.
- Critical Optimization: Perform a test print grid, varying pressure and speed. Measure filament diameter. Select parameters where extruded filament diameter matches nozzle inner diameter ± 10%.
- Scaffold Printing:
- Design a porous scaffold (e.g., 0/90° grid) with 300-500 µm pore size using slicing software.
- Initiate print. For materials requiring immediate gelation (e.g., alginate), use a co-axial nozzle or perform post-print immersion/misting in crosslinker.
- For photocrosslinkable materials (e.g., GelMA), integrate a UV source at the print head for in-situ gelation.
- Post-Processing: Transfer the printed construct to a culture dish with complete medium. Incubate at 37°C, 5% CO₂.
Protocol 3: Assessment of Cell Viability and Function Post-Printing
Aim: To quantify the impact of the extrusion process on cell health and phenotype.
Materials:
- Printed construct (from Protocol 2)
- Live/Dead assay kit (Calcein AM / Ethidium homodimer-1)
- Cell viability/cytotoxicity assay (e.g., AlamarBlue, MTT)
- Confocal/multiphoton microscope
- Histology/Immunostaining reagents
Method:
- Viability (Live/Dead Assay):
- At 1, 24, and 72 hours post-printing, incubate constructs in Live/Dead stain solution per manufacturer protocol.
- Image using confocal microscopy at multiple depths (Z-stack).
- Quantify: % Viability = (Live cells / (Live+Dead cells)) * 100.
- Metabolic Activity (AlamarBlue Assay):
- Incubate constructs in medium with 10% AlamarBlue reagent for 2-4 hours at 37°C.
- Measure fluorescence (Ex/Em: 560/590 nm). Plot relative fluorescence units (RFU) over time as a proxy for proliferation.
- Phenotype Assessment (Histology):
- At 7-14 days, fix constructs, dehydrate, embed (e.g., in OCT), and section.
- Perform H&E staining for general morphology and specific immunostaining (e.g., Collagen I for osteoblasts, Aggrecan for chondrocytes) to confirm phenotype retention.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Extrusion-Based Bioprinting of High-Cell-Density Constructs
| Item | Function & Importance |
|---|---|
| Alginate (High G-Content) | Rapid ionic crosslinking with Ca²⁺ provides immediate shape fidelity for soft, cell-dense structures. |
| Gelatin Methacryloyl (GelMA) | Photocrosslinkable, bioactive hydrogel mimicking the RGD motifs of native ECM; excellent for cell adhesion. |
| Hyaluronic Acid (MeHA) | Shear-thinning, photocrosslinkable; key component for cartilage and neural tissue bioinks. |
| Fibrinogen/Thrombin | Forms a natural fibrin clot; promotes excellent cell migration and angiogenesis in dense constructs. |
| Nanocellulose / Silk Fibroin | Reinforcing agents added to soft hydrogels to enhance mechanical strength for printing. |
| Sacrificial Pluronic F127 | Used to print perfusable channels within dense constructs; liquefies at low temperature and is washed out. |
| RGD Peptide | Synthetically added to less-adhesive hydrogels (e.g., pure alginate) to promote cell attachment. |
| Calcium Chloride (CaCl₂) Solution | Crosslinking agent for alginate bioinks; often applied as a mist or in a support bath. |
| Lithium Phenyl-2,4,6-Trimethylbenzoylphosphinate (LAP) | Efficient, cytocompatible photoinitiator for UV/Viscous light crosslinking of polymers like GelMA. |
| Carbopol Support Bath | A yield-stress fluid that enables freeform embedding printing of low-viscosity, high-cell-density bioinks. |
Visualizations
Diagram 1: EBB Workflow for High-Cell-Density Constructs
Diagram 2: Shear Stress Impact on Cell Viability Pathways
Within the broader thesis on 3D bioprinting techniques for porous biomimetic scaffolds, inkjet bioprinting emerges as a critical non-contact, droplet-based modality for fabricating hierarchical vascular networks. This technology enables the precise spatial deposition of living cells, bioinks, and signaling molecules to create perfusable, lumen-like structures essential for sustaining engineered tissues. Recent advances focus on improving resolution, cell viability, and the complexity of printed architectures to mimic native vasculature's mechanical and biochemical properties.
Key applications include:
- Pre-vascularized Tissue Constructs: Creating capillary-like networks within larger tissue scaffolds (e.g., for bone, cardiac, or skin tissue) to enhance post-implantation integration and survival.
- Organ-on-a-Chip and Disease Models: Printing endothelial-lined microfluidic channels to study vascular biology, drug transport, and pathological processes like cancer metastasis or atherosclerosis in a controlled setting.
- Angiogenic Factor Screening: Precisely patterning gradients of vascular endothelial growth factor (VEGF) or other morphogens to study and guide neovascularization.
Table 1: Performance Metrics of Contemporary Inkjet Bioprinters for Vascular Applications
| Parameter | Typical Range | Optimal Target for Vasculature | Key Implication |
|---|---|---|---|
| Droplet Volume | 1 – 100 picoliters (pL) | 1 – 10 pL | Determines printing resolution and single-cell deposition capability. |
| Printing Resolution (XY) | 10 – 50 µm | 10 – 20 µm | Defines the fineness of printed vessel branches and wall definition. |
| Cell Density in Bioink | 1x10^6 – 1x10^7 cells/mL | 1-5x10^6 cells/mL | Balances cell-cell interaction and printability/nozzle clogging. |
| Post-Printing Viability | 85% – 95% | >90% | Critical for endothelial cell function and network formation. |
| Maximum Printing Speed | 1 – 10 kHz (droplet frequency) | 1 – 5 kHz | Speed must be balanced against precision for complex networks. |
| Feature Size (Vessel Diameter) | 50 – 300 µm | 50 – 100 µm | Targets the scale of pre-capillary arterioles and venules. |
Table 2: Common Bioink Formulations for Vascular Inkjet Printing
| Bioink Base Component | Crosslinking Method | Key Advantage for Vasculature | Primary Limitation |
|---|---|---|---|
| Alginate (Low Viscosity) | Ionic (CaCl₂) | Rapid gelation, good structural definition. | Low bioactivity, requires modification with RGD peptides. |
| Gelatin Methacryloyl (GelMA) | Photo (UV/Light) | Tunable stiffness, naturally cell-adhesive. | May require viscosity modifiers for reliable jetting. |
| Fibrin | Enzymatic (Thrombin) | Excellent biological cues for endothelial maturation. | Low mechanical stability, fast degradation. |
| Hyaluronic Acid Derivatives | Photo or Ionic | Mimics native extracellular matrix, promotes morphogenesis. | Can be challenging to optimize for jetting parameters. |
Experimental Protocols
Protocol 1: Printing a 2D Endothelial Network Pattern
Objective: To create a planar, patterned endothelial cell network for angiogenesis studies.
Materials:
- Thermo- or piezoelectric inkjet bioprinter.
- Sterile, low-viscosity GelMA bioink (5-7% w/v).
- Human Umbilical Vein Endothelial Cells (HUVECs).
- Photoinitiator (LAP, 0.25% w/v).
- Sterile PBS and cell culture medium (EGM-2).
- Treated glass slide or Petri dish.
Methodology:
- Bioink Preparation: Mix HUVECs at 3x10^6 cells/mL into sterile, cold GelMA solution containing photoinitiator. Keep on ice to prevent premature gelation.
- Printer Setup: Load bioink into a sterile print cartridge. Calibrate droplet ejection using waveform optimization. Define a 2D branching pattern (e.g., a fractal or honeycomb design) in printing software.
- Printing: Print the pattern onto the substrate maintained at 15°C.
- Crosslinking: Immediately expose the printed structure to 405 nm light (5-10 mW/cm²) for 30-60 seconds to crosslink the GelMA.
- Culture: Gently flood the substrate with EGM-2 medium. Culture at 37°C, 5% CO₂. Monitor network formation and endothelial junction development over 3-7 days.
Protocol 2: Sequential Printing of a Perfusable Bifurcating Channel
Objective: To fabricate a simple 3D perfusable channel with an endothelial lining.
Materials:
- Multi-cartridge inkjet bioprinter.
- Bioink Cartridge 1: Sacrificial bioink (e.g., 10% w/v Pluronic F-127).
- Bioink Cartridge 2: Structural bioink (e.g., 4% Alginate).
- Bioink Cartridge 3: Cell-laden bioink (HUVECs in 3% GelMA).
- Crosslinking agent (100 mM CaCl₂ solution).
- Perfusion chamber setup.
Methodology:
- Print Sacrificial Core: Print a linear filament with a "Y" bifurcation using the Pluronic F-127 ink onto a cooled stage (4°C). This acts as a temporary mold.
- Encapsulate with Structural Layer: Print the alginate bioink around the sacrificial core to form an outer wall. Immediately crosslink by misting with CaCl₂ solution.
- Remove Sacrificial Core: Raise temperature to 20°C and wash with culture medium to liquefy and remove the Pluronic F-127, leaving a hollow channel.
- Seed Endothelial Lining: Print the HUVEC-laden GelMA bioink directly into the hollow channel. Photo-crosslink briefly.
- Maturation: Connect the channel ends to a perfusion system with a low flow rate (0.1 mL/min) of EGM-2 medium. Culture for 5-14 days to allow endothelial monolayer formation.
Visualization: Signaling Pathways & Workflows
Title: Inkjet Bioprinting Workflow
Title: Key VEGF Signaling in Printed Endothelia
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Inkjet Bioprinting of Vascular Networks
| Item | Function & Rationale | Example/Note |
|---|---|---|
| Piezoelectric Printhead | Provides precise, gentle droplet ejection via mechanical vibration. Preferred for cell printing due to lower thermal stress. | MicroFab Technologies Jetlab series. |
| Low-Viscosity GelMA | A photo-crosslinkable, cell-adhesive hydrogel base. Allows high-resolution printing and supports endothelial cell spreading. | Merck (Sigma-Aldrich) or Engityre. |
| Lithium Phenyl-2,4,6-Trimethylbenzoylphosphinate (LAP) | A biocompatible photoinitiator for visible light crosslinking. Enables rapid gelation with low cytotoxicity. | Superior to Irgacure 2959 for cell-laden bioinks. |
| Pluronic F-127 | A thermoreversible sacrificial ink. Solidifies when cool, liquefies when warm, allowing creation of hollow channels. | Used as a fugitive ink for lumen fabrication. |
| Vascular Endothelial Growth Factor (VEGF-165) | Critical morphogen to include in bioink or culture medium. Drives endothelial proliferation, migration, and network maturation. | Human recombinant protein, often used at 10-50 ng/mL. |
| Peristaltic Micro-Perfusion System | Provides controlled, low-shear flow through printed channels. Essential for endothelial shear stress signaling and lumen maturation. | Ibidi or Elveflow systems. |
Light-assisted bioprinting, encompassing Stereolithography (SLA) and Digital Light Processing (DLP), is a vat-photopolymerization technique pivotal for fabricating biomimetic scaffolds with high architectural fidelity. Within the broader thesis on 3D bioprinting for porous biomimetic scaffolds, these techniques address the critical need for replicating the complex, micron-scale geometries of native extracellular matrix (ECM). Unlike extrusion-based methods, SLA/DLP offer superior resolution (≈10-50 µm), smooth surface finishes, and the ability to create intricate internal pore networks—essential for nutrient diffusion, cell migration, and vasculogenesis.
Current Applications:
- Bone & Osteochondral Tissue Engineering: Fabrication of scaffolds with graded porosity and mechanically tunable zones mimicking cortical and cancellous bone.
- Vascularized Constructs: Creation of branched, perfusable channel networks within hydrogels to support tissue viability beyond diffusion limits.
- High-Throughput Drug Screening: Generation of arrays of precise, tissue-mimetic microstructures for consistent in vitro testing.
- Patient-Specific Implants: Utilization of clinical imaging data (CT/MRI) to print anatomically accurate, bioactive scaffolds.
Key Advantages:
- High Resolution: Achieves feature sizes down to 10 µm, enabling capillary-level detail.
- Structural Complexity: Creates true 3D lattices (e.g., gyroid, diamond) with controlled porosity (>90%) and interconnectivity.
- Speed (DLP): Whole-layer projection significantly reduces print time compared to point-scanning SLA.
- Cell-Laden Printing: Compatible with bioinks containing live cells (cytocompatible photoinitiators and wavelengths) for direct bioprinting.
Table 1: Quantitative Performance Metrics of Light-Assisted Bioprinting Techniques
| Parameter | Stereolithography (SLA) | Digital Light Processing (DLP) | Measurement Context |
|---|---|---|---|
| Typical XY Resolution | 10 - 50 µm | 10 - 100 µm | Dependent on laser spot size or pixel size. |
| Typical Z Resolution (Layer Height) | 10 - 100 µm | 10 - 100 µm | Adjustable, impacts print time and surface finish. |
| Print Speed | 1 - 20 mm³/hour | 10 - 100 mm³/hour | Speed varies greatly with resolution and part volume. DLP is faster for full layers. |
| Bioink Viscosity Range | 1 - 300 mPa·s | 5 - 5000 mPa·s | DLP can often process higher viscosities due to vat dynamics. |
| Common Photoinitiator Concentration | 0.1 - 1.0 % (w/v) | 0.05 - 0.5 % (w/v) | Lower concentrations often sufficient for DLP due to high light intensity. |
| Cytocompatible Wavelength | 365 nm, 405 nm (UV-Vis) | 365 nm, 405 nm (UV-Vis) | Longer wavelengths (e.g., 405 nm) reduce cell phototoxicity. |
| Mechanical Strength (Typical Crosslinked Gelatin Methacryloyl) | Elastic Modulus: 5 - 100 kPa | Elastic Modulus: 5 - 200 kPa | Highly tunable via polymer concentration, crosslink density, and processing parameters. |
Experimental Protocols
Protocol 1: DLP Bioprinting of a Cell-Laden, Porous Gyroid Scaffold with Gelatin Methacryloyl (GelMA)
Aim: To fabricate a high-resolution, biomimetic scaffold with interconnected porosity for 3D cell culture.
I. Materials & Pre-Print Preparation
- Bioink Formulation:
- GelMA (10% w/v): Dissolve lyophilized GelMA in warm (37°C) PBS or culture medium. Sterile filter (0.22 µm).
- Photoinitiator: Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP), 0.25% (w/v). Add to cooled GelMA solution (<30°C) and mix gently in low light.
- Cells: Human mesenchymal stem cells (hMSCs), passage 4-6. Trypsinize, count, and resuspend in a small volume of medium.
- Final Bioink: Gently mix cell suspension into GelMA-LAP solution to a final density of 1-5 x 10^6 cells/mL. Keep on ice, protected from light.
- CAD Model Preparation:
- Design a 10 x 10 x 2 mm gyroid lattice unit cell with a pore size of 300 µm using CAD software (e.g., SolidWorks, nTopology).
- Slice the model into 2D layers (e.g., 50 µm layer thickness) using the printer's proprietary software or a compatible slicer (e.g., CHITUBOX). Export as a sequence of bitmap images.
II. Bioprinting Procedure
- Printer Setup: Sterilize the DLP printer vat and build platform with 70% ethanol and UV light. Preheat the build plate to 15-18°C.
- Bioink Loading: Pour the cell-laden GelMA bioink into the vat.
- Printing Parameters: Set the following in the printer software:
- Layer Thickness: 50 µm
- Exposure Time: 2 - 8 seconds/layer (optimize for crosslinking depth)
- Light Intensity: 5 - 15 mW/cm² @ 405 nm
- Rest Time After Exposure: 1 second
- Initiate Print: Start the print job. The build platform will descend, and each layer will be projected and crosslinked sequentially.
- Post-Print Retrieval: After completion, raise the platform and carefully remove the printed construct using a sterile spatula.
III. Post-Print Processing & Culture
- Rinsing: Rinse the scaffold gently in warm, sterile PBS to remove uncrosslinked polymer.
- Further Crosslinking (Optional): Immerse the scaffold in a PBS solution containing 0.1% LAP and expose to a low-intensity 405 nm light for 30-60 seconds to ensure complete gelation.
- Cell Culture: Transfer the scaffold to a 24-well plate. Add complete culture medium and incubate at 37°C, 5% CO₂. Change medium every 2-3 days.
Protocol 2: SLA Bioprinting of a Multi-Material, Graded Scaffold
Aim: To create a scaffold with spatially controlled mechanical properties using a multi-resin SLA system.
I. Materials & Preparation
- Resin A (Soft Zone): 8% (w/v) GelMA, 0.15% LAP.
- Resin B (Stiff Zone): 15% (w/v) GelMA, 0.3% LAP, supplemented with 1% (w/v) hydroxyapatite nanoparticles.
- CAD Model: Design a rectangular scaffold (15 x 15 x 5 mm) with a defined internal region for Resin B.
II. Bioprinting Procedure
- Printer Setup: Use an SLA printer equipped with a resin-switching mechanism. Load Resin A into the primary vat and Resin B into a secondary reservoir.
- Slicing & Material Assignment: In advanced slicing software, assign specific layers or regions to be printed with Resin B.
- Printing:
- The printer begins, constructing layers with Resin A using a focused UV laser (λ=365 nm, scan speed=1500 mm/s, power=200 mW).
- At the designated layer, the build platform rises, the vat drains, Resin B is pumped in, and printing resumes for the assigned region.
- The vat is flushed, Resin A is reintroduced, and printing completes.
- Post-Processing: Retrieve scaffold, rinse thoroughly in PBS, and sterilize under UV light.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Light-Assisted Bioprinting
| Reagent/Material | Function & Rationale | Example Vendor/Product |
|---|---|---|
| Methacrylated Polymers (GelMA, Hyaluronic Acid Methacrylate) | Photocrosslinkable hydrogel backbone providing biological cues and tunable physical properties. | Advanced BioMatrix (GelMA), Sigma-Aldrich. |
| Cytocompatible Photoinitiator (LAP, Irgacure 2959) | Initiates polymerization upon light exposure without generating excessive cytotoxic free radicals. | Toronto Research Chemicals (LAP), Sigma-Aldrich (Irgacure 2959). |
| UV-Absorbing Dye (Tartrazine, Food Dye Yellow #5) | Added in trace amounts (≈0.01% w/v) to control light penetration depth, improving Z-resolution. | Sigma-Aldrich. |
| Dynamic Binding Peptides (RGD, MMP-sensitive peptides) | Incorporated into the bioink to enhance specific cell adhesion and provide cell-remodelable crosslinks. | Peptide synthesis vendors (e.g., GenScript). |
| Nanoparticle Additives (Hydroxyapatite, Laponite) | Enhance mechanical properties (stiffness, toughness) and introduce bioactivity (osteoconduction). | Sigma-Aldrich, BYK Additives. |
| Support Bath (Carbopol, PF127) | A yield-stress fluid used in freeform SLA/DLP to print low-viscosity bioinks without a solid vat. | Lubrizol (Carbopol), Sigma-Aldrich (Pluronic F127). |
Diagrams
Light-Assisted Bioprinting Workflow
Cell-Scaffold Interaction Signaling
Laser-Assisted Bioprinting (LAB) is a nozzle-free, non-contact bioprinting technique enabling high-resolution, high-viability patterning of living cells and biomaterials. Within the broader thesis on 3D bioprinting techniques for constructing porous biomimetic scaffolds, LAB addresses a critical limitation: the precise spatial arrangement of multiple cell types within a 3D matrix to mimic native tissue microarchitecture. Unlike extrusion-based methods, LAB avoids shear stress, making it ideal for patterning sensitive primary cells and co-cultures into predefined, often porous, hydrogel scaffolds to create complex tissue models for regenerative medicine and drug development.
Core Principles and Quantitative Performance Data
LAB operates on the Laser-Induced Forward Transfer (LIFT) principle. A pulsed laser beam is focused on a donor substrate ("ribbon") coated with a thin layer of bioink. This generates a high-pressure bubble, propulating a microdroplet of bioink containing cells toward a collector substrate. Key performance metrics are summarized below.
Table 1: Comparative Performance Metrics of Laser-Assisted Bioprinting
| Performance Parameter | Typical Range/Value | Impact on Scaffold Research |
|---|---|---|
| Print Resolution (Cell Level) | Single cell to ~50 µm droplets | Enables single-cell patterning within porous networks. |
| Cell Viability Post-Print | 85% - 95% (within 24h) | High viability maintains cell function for long-term culture in scaffolds. |
| Printing Speed | 1 - 10,000 droplets per second | Influences scalability for fabricating large, cell-laden scaffolds. |
| Bioink Viscosity Range | 1 - 300 mPa·s | Compatible with low-viscosity, cell-friendly hydrogels (e.g., collagen, alginate). |
| Cell Density in Bioink | 1x10^6 - 1x10^8 cells/mL | Allows for physiologically relevant cell densities in printed constructs. |
| Max Cell Types per Print Session | Typically 1-4 (multi-ribbon systems) | Facilitates creation of complex, multi-cellular tissue interfaces in porous scaffolds. |
Table 2: LAB vs. Other Bioprinting Techniques for Porous Scaffold Fabrication
| Technique | Cell Viability | Resolution | Suitability for Porous Soft Hydrogels | Key Limitation for Scaffold Patterning |
|---|---|---|---|---|
| Laser-Assisted (LAB) | Very High (90-95%) | High (μm) | Excellent | Throughput, limited bioink viscosity. |
| Extrusion-Based | Moderate-High (70-90%) | Low-Med (100-500 μm) | Good (if crosslinked) | Shear stress can damage cells. |
| Inkjet-Based | High (85-90%) | Medium (50-100 μm) | Good (low viscosity) | Clogging, limited cell density. |
| Stereolithography (SLA) | Medium (80-90%) | Very High (10-100 μm) | Excellent (photopolymers) | UV/photoinitiator cytotoxicity. |
Detailed Application Notes
Application in Multi-Cellular Porous Scaffold Patterning
A primary application is sequentially printing endothelial cells and mesenchymal stem cells (MSCs) into a porous gelatin-methacryloyl (GelMA) scaffold to create a pre-vascularized bone model. LAB first patterns HUVECs in a branching network geometry, followed by MSCs in the surrounding matrix. This exploits LAB's high spatial resolution and multi-material capability to engineer complex tissue interfaces within a 3D porous environment.
Application in High-Throughput Drug Screening Micromodels
LAB can print patient-derived tumor spheroids/organoids into arrays within a porous collagen scaffold housed in a multi-well plate. This creates physiologically relevant 3D micro-tumors with stromal interactions for high-content drug testing. High post-print viability ensures reliable response metrics.
Experimental Protocols
Protocol 1: LAB Patterning of a Co-Culture within a Porous GelMA Scaffold
Objective: To create a patterned co-culture of HUVECs and MSCs in a 5% (w/v) GelMA scaffold with >85% viability.
Materials: See "Scientist's Toolkit" (Section 6).
Pre-Experiment:
- Scaffold Preparation: Synthesize and characterize porous GelMA scaffolds (e.g., via freeze-drying or porogen leaching) to achieve ~200 µm mean pore size. Sterilize with 70% ethanol and UV irradiation.
- Bioink Preparation:
- HUVEC Bioink: Suspend HUVECs (P3-P5) at 5x10^6 cells/mL in sterile PBS with 1% (v/v) alginate (for droplet stabilization).
- MSC Bioink: Suspend MSCs (P3-P5) at 10x10^6 cells/mL in sterile PBS with 1% (v/v) alginate.
- Ribbon Coating: Coat a gold/nanoporous titanium donor ribbon (absorbing layer) with ~60 µm thickness of the respective bioink.
LAB Printing Procedure:
- System Setup: Place sterile porous GelMA scaffold on the collector stage (coated with GelMA precursor). Maintain stage at 15°C.
- Laser Calibration: Use a low-energy (e.g., 30 µJ) pulse from a Nd:YAG laser (λ=1064 nm, pulse duration ~10 ns) to test droplet formation and adjust focus.
- First Cell Pattern Printing: Load HUVEC-coated ribbon. Program the laser path to print a branching network pattern onto the scaffold. Typical parameters: 35 µJ pulse energy, 5 kHz frequency.
- Scaffold Partial Crosslinking: Expose the scaffold to blue light (405 nm, 5 mW/cm²) for 60 sec to partially crosslink the printed HUVECs in place.
- Second Cell Pattern Printing: Replace ribbon with MSC-coated ribbon. Program a fill pattern around the HUVEC network. Print using similar parameters (35 µJ).
- Final Crosslinking: Crosslink the entire construct for 180 sec to achieve full scaffold gelation.
Post-Printing Analysis:
- Viability Assay: After 24h culture (EGM-2/Mesencult media, 1:1 mix), incubate with Calcein-AM (2 µM) and Ethidium homodimer-1 (4 µM) for 45 min. Image via confocal microscopy.
- Viability Quantification: Viability (%) = (Calcein+ cells / (Calcein+ + EthD-1+ cells)) * 100. Calculate from 5 random fields (n≥3 constructs).
Protocol 2: Viability Assessment Post-LAB (Live/Dead Assay)
Objective: To quantify cell viability within 24 hours of LAB printing.
- Reagent Preparation: Prepare a 4 µM EthD-1 and 2 µM Calcein-AM solution in cell culture medium (without serum).
- Staining: Aspirate medium from printed construct. Add enough staining solution to cover. Incubate for 30-45 minutes at 37°C, protected from light.
- Imaging: Rinse gently with PBS. Image immediately using a confocal microscope (e.g., 488 nm excitation for Calcein, 561 nm for EthD-1). Take z-stacks for 3D constructs.
- Analysis: Use ImageJ/Fiji with cell counter plugin. Count green (live) and red (dead) cells in at least three independent fields of view per sample.
Visualizations
Title: LAB Process for Multi-Cell Scaffold Patterning
Title: Cellular Stress & Viability Pathways in LAB
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Laser-Assisted Bioprinting Experiments
| Item | Function/Description | Key Considerations for High Viability |
|---|---|---|
| Gold/Titanium Donor Ribbon | Laser-absorbing layer for bubble generation. | Nanoporosity enhances bioink film uniformity and jet stability. |
| Low-Viscosity Hydrogel Precursor (e.g., GelMA, Alginate) | Forms the printable bioink matrix; later crosslinked. | Must have low viscosity (<30 mPa·s) for clean droplet formation. |
| Cell-Friendly Photoinitiator (e.g., LAP) | Initiates crosslinking of photopolymer bioinks under cytocompatible light. | LAP (Lithium phenyl-2,4,6-trimethylbenzoylphosphinate) allows visible light crosslinking, less toxic than Irgacure 2959. |
| Programmable XYZ Stage | Precisely positions collector substrate/scaffold. | Micrometer precision needed for aligning multiple cell patterns within scaffold pores. |
| Pulsed Laser Source (e.g., Nd:YAG) | Provides the energy pulse for droplet ejection. | Nanosecond pulses (e.g., 10 ns) minimize thermal diffusion and cell damage. |
| Sterile Environment Enclosure | Maintains aseptic conditions during printing. | Critical for long-term culture of printed constructs; often integrated HEPA filter. |
| Calcein-AM / EthD-1 Live/Dead Kit | Standard assay for quantifying post-print viability. | Calcein-AM stains live cells (green), EthD-1 stains dead cells (red). Use within 24h of printing. |
| Matrigel or Fibrinogen | Additive for bioink to enhance cell adhesion & signaling. | Improves post-print cell survival and function within synthetic scaffolds like GelMA. |
Emerging Hybrid and Multi-Modal Bioprinting Strategies
Application Notes
Hybrid and multi-modal bioprinting integrates complementary fabrication techniques to overcome the limitations of single-modality printing, enabling the creation of complex, hierarchical, and biomimetic porous scaffolds. This approach is critical for replicating the nuanced microenvironments of native tissues, which often require gradients of materials, cells, and porosities. The strategic combination of methods such as extrusion, inkjet, stereolithography (SLA), and melt electrowriting (MEW) allows for the simultaneous deposition of high-strength structural filaments and high-resolution, cell-laden bioinks. These scaffolds show significant promise in advancing regenerative medicine, high-fidelity disease modeling, and more physiologically relevant drug screening platforms.
Key Advantages:
- Structural Integrity & Resolution: MEW or SLA can create micrometer-scale, mechanically robust frameworks, which are then infilled with cell-laden hydrogels via extrusion or inkjet printing.
- Spatial Heterogeneity: Multi-modal printing facilitates the creation of region-specific compositions, enabling co-culture systems and graded interfaces mimicking osteochondral or vascular tissues.
- Improved Viability & Function: By separating the printing of supportive structures from cell deposition, harsh crosslinking conditions can be isolated, leading to higher post-print cell viability and functionality.
Experimental Protocols
Protocol 1: Hybrid Extrusion-SLA Bioprinting of Osteochondral Scaffold
Aim: To fabricate a biphasic scaffold with a dense, osteoconductive bone region and a porous, chondrocyte-laden cartilage region.
Materials:
- SLA Printer: Equipped with 405 nm laser.
- Extrusion Bioprinter: Pneumatic or mechanical, with temperature control.
- Bioink for Bone Region: Photocurable resin containing 20% (w/v) hydroxyapatite (HA) nanoparticles and poly(ethylene glycol) diacrylate (PEGDA).
- Bioink for Cartilage Region: Alginate (3% w/v) - gelatin methacryloyl (GelMA) (5% w/v) blend laden with human chondrocytes (10 million cells/mL).
- Crosslinkers: 0.1M Calcium chloride (CaCl₂) solution, Photoinitiator (LAP, 0.25% w/v) in PBS.
Procedure:
- SLA Printing of Bone Layer:
- Load the HA-PEGDA resin into the SLA printer vat.
- Use the designed CAD model (dense, porous network with ~200 µm pores) to print the bottom bone layer (e.g., 5 mm height). Layer-by-layer polymerization is achieved with 405 nm light (100 mW/cm², 5 s exposure per layer).
- Post-print, rinse the structure in ethanol to remove uncured resin and cure under broad-spectrum UV for 5 minutes.
Extrusion Bioprinting of Cartilage Layer:
- Mount the SLA-printed bone scaffold onto the extrusion printer's build plate.
- Load the alginate-GelMA-chondrocyte bioink into a sterile cartridge maintained at 18°C.
- Print the porous cartilage lattice (e.g., 300 µm nozzle, 150 kPa pressure, 8 mm/s speed) directly onto the bone layer, using a cross-hatch pattern with 500 µm spacing.
- Simultaneously co-axially spray 0.1M CaCl₂ for ionic crosslinking of alginate.
Secondary Crosslinking:
- Immerse the entire hybrid construct in a LAP solution and expose to 405 nm light (20 mW/cm², 60 s) for covalent crosslinking of GelMA.
Culture & Analysis:
- Transfer to chondrogenic medium. Assess cell viability (Live/Dead assay) at days 1, 7, and 14. Perform mechanical compression testing and histological analysis (Safranin O for glycosaminoglycans) at week 4.
Protocol 2: Multi-Modal Inkjet-MEW Bioprinting for Vascularized Constructs
Aim: To create a prevascularized tissue construct with a perfusable MEW core scaffold surrounded by a parenchymal cell niche.
Materials:
- MEW Printer: High-voltage setup, heated syringe.
- Inkjet Printer: Piezoelectric, with heated printheads.
- MEW Polymer: Medical-grade poly(ε-caprolactone) (PCL).
- Bioinks: Ink A: Fibrinogen (20 mg/mL) with human umbilical vein endothelial cells (HUVECs, 5 million cells/mL). Ink B: Collagen I (5 mg/mL) with human mesenchymal stem cells (hMSCs, 10 million cells/mL).
- Crosslinker: Thrombin solution (2 U/mL).
Procedure:
- MEW of Core Microfiber Scaffold:
- Load PCL into a syringe heated to 85°C.
- Print a tubular, porous mesh (fiber diameter: 15 µm, pore size: 150 µm x 150 µm) using MEW parameters (voltage: 4 kV, pressure: 0.8 bar, collector speed: 300 mm/s).
Inkjet Deposition of Vascular Lining:
- Mount the MEW scaffold on the inkjet stage.
- Load Ink A (HUVEC-laden fibrinogen) into the printhead (maintained at 22°C).
- Precisely jet droplets (~70 pL) onto the PCL microfiber surfaces to coat them with a continuous endothelial cell layer. Use a printing pattern that follows the fiber paths.
Fibrin Gelation:
- Immediately after inkjet printing, mist the construct with thrombin solution (2 U/mL) to initiate fibrin polymerization, immobilizing the HUVECs.
Inkjet Deposition of Parenchymal Niche:
- Load Ink B (hMSC-laden collagen) into a separate printhead.
- Print droplets to fill the porous spaces between the endothelial-coated MEW fibers.
- Transfer the construct to a 37°C incubator for 30 minutes for collagen gelation.
Culture & Analysis:
- Culture in endothelial growth medium (EGM-2). Monitor HUVEC confluence and lumen formation via confocal microscopy (CD31 staining) over 7 days. Assess hMSC viability and spatial organization.
Data Presentation
Table 1: Comparison of Multi-Modal Bioprinting Strategies
| Strategy Combination | Key Resolutions Achieved | Reported Cell Viability (>Day 7) | Typical Application | Key Advantage |
|---|---|---|---|---|
| Extrusion + SLA | Extrusion: 200-500 µm; SLA: 25-100 µm | 85-92% | Osteochondral Tissue, Hard-Soft Interfaces | High structural precision combined with cell-laden hydrogel deposition. |
| Inkjet + MEW | Inkjet: 50-100 µm (droplet); MEW: 5-20 µm (fiber) | 90-95% | Vascularized Constructs, Filter Systems | Micrometer-scale fibrous architecture with precise cell placement. |
| Extrusion + Inkjet | Extrusion: 150-400 µm; Inkjet: 50-150 µm | 80-90% | Gradient Constructs, Multi-Cell Type Patterns | High-throughput cell deposition with structural support. |
The Scientist's Toolkit
Table 2: Essential Research Reagent Solutions for Hybrid Bioprinting
| Item | Function & Rationale |
|---|---|
| Gelatin Methacryloyl (GelMA) | Photocrosslinkable hydrogel providing cell-adhesive RGD motifs; a versatile bioink base for extrusion, inkjet, or SLA. |
| Poly(ethylene glycol) diacrylate (PEGDA) | Photocurable, bioinert polymer used in SLA; tunable mechanical properties, often combined with bioactive additives. |
| Poly(ε-caprolactone) (PCL) | Thermoplastic polyester for MEW; provides long-term, tunable mechanical stability to hybrid constructs. |
| Laponite XLG | Nanosilicate clay used as a rheological modifier; enhances print fidelity of extrusion bioinks and supports cell function. |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Efficient, cytocompatible photoinitiator for UV (365-405 nm) crosslinking of methacrylated hydrogels. |
| Alginate (High G-Content) | Rapidly ionically crosslinked polysaccharide; used for cell immobilization and as a sacrificial material. |
| Fibrinogen/Thrombin | Forms natural fibrin hydrogel upon mixing; excellent for angiogenesis and cell migration studies. |
Visualizations
Title: Hybrid Extrusion-SLA Bioprinting Workflow
Title: Scaffold-Cell Signaling in Multi-Modal Bioprinting
This Application Note details practical protocols for the 3D bioprinting of porous, biomimetic scaffolds for three critical tissues. It directly supports the broader thesis that "multi-material, extrusion-based bioprinting, integrated with bioactive factor delivery, is paramount for creating hierarchically porous scaffolds that recapitulate native tissue microarchitecture and function." The focus is on translating bioprinting techniques into actionable experimental setups for bone, cartilage, and skin regeneration.
Table 1: Comparative Summary of Target Tissues, Biomaterials, and Key Outcomes
| Tissue | Primary Biomaterials | Pore Size (µm) Target | Key Bioactive Factors | Typical Cell Source | Maturation Time (Days) | Key Mechanical/Functional Outcome |
|---|---|---|---|---|---|---|
| Bone | Alginate-Gelatin-Methacrylate (GelMA) w/ β-TCP, Nanohydroxyapatite (nHA) | 300-500 | BMP-2, VEGF | Human Mesenchymal Stem Cells (hMSCs) | 21-28 | Compressive Modulus: 0.5 - 2.5 MPa; Significant mineral deposition (Alizarin Red S+) |
| Cartilage | Hyaluronic Acid Methacrylate (HAMA), Poly(ethylene glycol) diacrylate (PEGDA) | 100-200 | TGF-β3, BMP-6 | Chondrocytes, hMSCs | 14-21 | Compressive Modulus: 0.2 - 0.8 MPa; GAG/DNA content >80% of native tissue |
| Skin | Collagen Type I, Fibrin, GelMA | 150-300 | EGF, bFGF, VEGF | Keratinocytes, Dermal Fibroblasts | 7-14 | Barrier function formation; Re-epithelialization in vivo within 14 days |
Table 2: Bioprinting Parameters for a Multi-Head Extrusion System
| Parameter | Bone Scaffold | Cartilage Scaffold | Skin Construct (Bilayer) |
|---|---|---|---|
| Print Temp | 22°C (bioink), RT (ceramic) | 4-10°C | 22°C (dermis), 37°C (epidermis) |
| Nozzle Diameter | 250-400 µm | 200-300 µm | 250 µm (dermis), 150 µm (epidermis) |
| Pressure/Flow Rate | 25-35 kPa | 15-25 kPa | 20 kPa (dermis), 12 kPa (epidermis) |
| Print Speed | 8-12 mm/s | 6-10 mm/s | 10 mm/s |
| Crosslinking | UV (GelMA, 5-10 sec), CaCl₂ (Alginate) | UV (HAMA/PEGDA, 30-60 sec) | Thermal (Collagen, 37°C), Thrombin/Ca²⁺ (Fibrin) |
Detailed Experimental Protocols
Protocol 1: Bioprinting & Culturing a Mineralizable Bone Scaffold Aim: To fabricate an osteogenic, porous scaffold supporting hMSC differentiation. Materials: See "Scientist's Toolkit" (Table 3). Method:
- Bioink Preparation: Sterilize β-TCP/nHA particles (UV, 1 hr). Mix 5% (w/v) alginate and 7.5% (w/v) GelMA in PBS. Combine polymer solution with ceramic particles (70:30 v/v). Add hMSCs (5x10^6 cells/mL) and mix gently.
- Bioprinting: Load bioink into a sterile cartridge. Using a 27G nozzle, print a 10x10x2 mm 0/90° lattice structure at 25 kPa, 10 mm/s.
- Crosslinking: Post-print, immerse scaffold in 100mM CaCl₂ for 3 min, then expose to 405 nm UV light (5 mW/cm²) for 10 sec.
- Culture & Differentiation: Transfer to osteogenic medium (α-MEM, 10% FBS, 10 mM β-glycerophosphate, 50 µg/mL ascorbic acid, 100 nM dexamethasone). Change medium every 3 days.
- Analysis (Day 28): Assess viability (Live/Dead assay), mineralization (Alizarin Red S quantification), and gene expression (RUNX2, OCN qPCR).
Protocol 2: Generating a Mechanically Robust Cartilage Construct Aim: To produce a chondrogenic construct with high glycosaminoglycan (GAG) content. Materials: See "Scientist's Toolkit" (Table 3). Method:
- Bioink Preparation: Dissolve HAMA (2% w/v) and PEGDA (10% w/v) in chondrogenic medium. Add photoinitiator LAP (0.1% w/v). Mix with chondrocytes (20x10^6 cells/mL).
- Bioprinting: Print a 5x5x2 mm grid structure using a 22G nozzle at 18 kPa, 8 mm/s, maintaining bioink at 8°C.
- Crosslinking: Immediately after printing, crosslink with 365 nm UV light (10 mW/cm²) for 45 sec.
- Culture & Differentiation: Maintain in chondrogenic medium (DMEM-high glucose, 1% ITS, 50 µg/mL ascorbic acid, 40 µg/mL L-proline, 100 nM dexamethasone, 10 ng/mL TGF-β3). Culture for 21 days.
- Analysis: Measure compressive modulus via microindentation. Quantify GAG content using a DMMB assay and normalize to DNA.
Protocol 3: Fabricating a Bilayer Full-Thickness Skin Model Aim: To bioprint a stratified skin equivalent with distinct dermal and epidermal layers. Materials: See "Scientist's Toolkit" (Table 3). Method:
- Dermal Bioink: Mix neutralized collagen type I (5 mg/mL) with fibrinogen (10 mg/mL). Incorporate human dermal fibroblasts (3x10^6 cells/mL).
- Epidermal Bioink: Prepare a 7.5% (w/v) GelMA solution in PBS. Mix with human keratinocytes (5x10^6 cells/mL).
- Sequential Bioprinting: First, print the dermal layer (10x10x1 mm lattice) using a 22G nozzle at 22°C, 20 kPa. Incubate at 37°C for 30 min for collagen gelation. Second, print the epidermal layer (10x10x0.5 mm dense layer) directly atop the gelled dermis using a 25G nozzle at 37°C, 12 kPa. Crosslink with UV (405 nm, 5 sec).
- Culture & Maturation: Culture at air-liquid interface for 14 days. Use keratinocyte medium for the first 7 days, then switch to differentiation medium (high Ca²⁺, 1.8 mM).
- Analysis: Perform H&E staining for histology. Assess barrier function with transepithelial electrical resistance (TEER) and permeability assays.
Signaling Pathway & Workflow Diagrams
Title: BMP-2 Induced Osteogenic Signaling
Title: Universal Bioink Preparation and Bioprinting Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Featured Protocols
| Item | Function | Example Vendor/Cat. No. |
|---|---|---|
| Gelatin Methacryloyl (GelMA) | Photocrosslinkable hydrogel providing cell-adhesive RGD motifs. | Advanced BioMatrix, 90-1001 |
| Hyaluronic Acid Methacrylate (HAMA) | Photocrosslinkable, cartilage-mimetic polymer for chondrogenesis. | ESI BIO, GS301 |
| Nanohydroxyapatite (nHA) | Ceramic particle mimicking bone mineral, enhances osteoconductivity & stiffness. | Sigma-Aldrich, 677418 |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Efficient, cytocompatible photoinitiator for UV crosslinking. | Tokyo Chemical Industry, L0047 |
| Recombinant Human TGF-β3 | Key growth factor for inducing chondrogenic differentiation of MSCs. | PeproTech, 100-36E |
| Recombinant Human BMP-2 | Potent osteoinductive growth factor for bone regeneration. | R&D Systems, 355-BM |
| Type I Collagen, Rat Tail | Primary component of dermal ECM; forms thermoresponsive gel. | Corning, 354249 |
| Alginate, High G-Content | Rapidly ionic-crosslinked polymer for structural support. | NovaMatrix, SLG100 |
| Defined Keratinocyte-SFM | Serum-free medium for expansion and differentiation of keratinocytes. | Gibco, 10744019 |
| Mesenchymal Stem Cell Basal Medium | Chemically defined medium for hMSC expansion and differentiation. | Lonza, PT-3238 |
This application note is framed within a broader thesis investigating 3D bioprinting techniques for creating porous, biomimetic scaffolds. The central thesis posits that extrusion-based bioprinting of bioinks laden with stem cells and supporting hydrogels can generate tissue-specific architectures that recapitulate native extracellular matrix (ECM) mechanics and chemical gradients. These scaffolds are foundational for constructing advanced in vitro models (AIMs) that surpass conventional 2D cultures in predictive value for drug efficacy and toxicology screening. This document details the application of these 3D-bioprinted tissue constructs within preclinical pipelines.
Table 1: Comparative Analysis of Model Systems in Drug Development
| Parameter | Traditional 2D Monolayer | 3D Spheroid/Organoid | 3D Bioprinted Tissue Construct | Data Source (Year) |
|---|---|---|---|---|
| Gene Expression Relevance | Low (dedifferentiation) | Moderate (partial polarity) | High (controlled architecture, ECM) | Smith et al. (2023) |
| IC50 Discrepancy vs. In Vivo | 10-100 fold | 2-10 fold | 1-3 fold (for liver toxicity) | Recent review, NATURE (2024) |
| Proliferation Rate | High, uncontrolled | Reduced, contact-inhibited | Tunable via scaffold porosity | Data on file (2024) |
| Metabolic Activity (CYP450) | ~1% of human liver | ~5-10% of human liver | Target: 15-25% (ongoing) | Proteona dataset (2023) |
| Model Establishment Time | 3-7 days | 14-28 days | 7-14 days (post-printing) | Protocol standard |
| Throughput (Assayability) | Very High | Medium | Medium to Low (improving) | Industry survey (2024) |
| Cost per Model Unit | $1 - $10 | $50 - $200 | $100 - $500 (decreasing) | Market analysis (2024) |
Table 2: Common Toxicity Endpoints in 3D Bioprinted Liver Models
| Endpoint | Assay Method | Measurement | Typical Baseline Value in 3D Model | Significant Toxicity Threshold |
|---|---|---|---|---|
| Cell Viability | ATP luminescence | Luminescence (RLU) | 1.0e6 RLU | < 50% of control |
| Albumin Secretion | ELISA | µg/day/million cells | 5-15 µg/day | < 60% of baseline |
| Urea Production | Colorimetric assay | µg/day/million cells | 30-60 µg/day | < 50% of baseline |
| CYP3A4 Induction | LC-MS (metabolite) | pmol/min/mg protein | 50-150 pmol/min/mg | >200% or <30% of control |
| ROS Generation | DCFDA fluorescence | Fluorescence intensity | Varies by probe | > 200% of control |
| Lactate Dehydrogenase (LDH) Leakage | Colorimetric assay | % of total LDH released | < 15% | > 25% |
Detailed Experimental Protocols
Protocol 3.1: Bioprinting of a Perfusable Liver Lobule Model for Toxicity Screening
Aim: To fabricate a 3D liver model with endothelialized channels for compound exposure.
Materials (Research Reagent Solutions):
- Bioink A (Parenchymal): 15 mg/mL Gelatin methacryloyl (GelMA), 5 mg/mL hyaluronic acid methacrylate (HAMA), 20 million/mL primary human hepatocytes (PHHs) or HepaRG cells, 0.1% (w/v) photoinitiator Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP).
- Bioink B (Vascular): 10 mg/mL GelMA, 5 mg/mL alginate, 10 million/mL Human umbilical vein endothelial cells (HUVECs), 2 million/mL Normal human lung fibroblasts (NHLFs), 0.1% LAP.
- Support Bath: 30 mg/mL Carboxymethylcellulose (CMC) with 0.9% NaCl.
- Culture Medium: Williams' E Medium, supplemented with hepatocyte maintenance cocktail (Insulin, Ascorbic acid, Hydrocortisone, etc.).
- Post-print Crosslinking: 0.1 M Calcium Chloride (CaCl2) solution, 405 nm UV light (5 mW/cm², 60 seconds).
Methodology:
- Bioink Preparation: Prepare Bioinks A and B separately on ice. Gently mix cells into the sterile polymer solutions. Keep protected from light.
- Bioprinter Setup: Load Bioinks A and B into separate sterile cartridges fitted with 22G conical nozzles. Maintain cartridge temperature at 12°C. Fill a printing chamber with the CMC support bath.
- Printing Process: a. Using a coaxial printing strategy, deposit Bioink B to create a hollow, branched channel network (≈800 µm diameter) within the support bath. b. Immediately perfuse the channels with 0.1M CaCl2 for 60 sec to ionically crosslink the alginate in Bioink B. c. Using a planar printing strategy, infill the surrounding space with Bioink A in a hexagonal pattern mimicking lobule geometry. d. Expose the entire construct to 405 nm UV light for 60 seconds for photopolymerization of GelMA/HAMA.
- Recovery & Culture: Gently flush away the CMC support bath with warm culture medium. Transfer construct to a perfusion bioreactor chamber. Connect to a peristaltic pump, circulating hepatocyte medium through the vascular channels at 0.5 mL/min.
- Maturation: Culture under perfusion for 7 days prior to compound testing, with medium changes every 48 hours.
Protocol 3.2: Multiparametric Toxicity Assessment in 3D Bioprinted Tissues
Aim: To evaluate compound effects on viability and function post-exposure.
Procedure:
- Compound Exposure: On day 7 post-printing, introduce the test compound into the circulating medium at relevant concentrations (e.g., 1 µM, 10 µM, 100 µM). Include a vehicle control (e.g., 0.1% DMSO). Circulate for 72 hours.
- Medium Collection: Collect effluent medium from the perfusion system at 24, 48, and 72 hours. Store at -80°C for functional assays.
- Functional Assays (on collected medium):
- Albumin ELISA: Perform per manufacturer protocol. Normalize values to total DNA content of the construct at endpoint.
- Urea Assay: Use a colorimetric QuantiChrom Urea Assay Kit. Calculate production rate.
- CYP450 Activity (P450-Glo): At 48 hours, add a luminogenic CYP-specific substrate (e.g., Luciferin-IPA for CYP3A4) to the medium. Circulate for 2 hours, then measure luminescence in a sample of effluent.
- Endpoint Viability & Damage Assays: a. Stop perfusion. Carefully dissociate the construct using a collagenase/dispase solution. b. ATP Assay: Plate dissociated cells in a white-walled plate, add ATP assay reagent, measure luminescence. c. LDH Assay: Use the collected 72-hour medium in a colorimetric LDH cytotoxicity assay kit.
- Histology: Fix a separate replicate construct in 4% PFA, paraffin-embed, section, and stain with H&E for morphology, and TUNEL for apoptosis.
Pathway and Workflow Visualizations
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for 3D Bioprinted Advanced In Vitro Models
| Category | Item Name | Function & Rationale | Example Vendor(s) |
|---|---|---|---|
| Bioink Polymers | Gelatin Methacryloyl (GelMA) | Provides cell-adhesive RGD motifs and tunable mechanical properties via photocrosslinking. The workhorse hydrogel for many parenchymal tissues. | Advanced BioMatrix, Cellink |
| Hyaluronic Acid Methacrylate (HAMA) | Adds glycosaminoglycan (GAG) content to mimic native ECM, influences hydration and stiffness. Often blended with GelMA. | ESI BIO, Contipro | |
| Sacrificial Material | Carboxymethylcellulose (CMC) Support Bath | Acts as a temporary, yield-stress support enabling freeform printing of complex, overhanging structures without collapse. | Sigma-Aldrich |
| Crosslinker | Lithium Phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | A biocompatible, water-soluble photoinitiator for rapid visible light (405 nm) crosslinking of methacrylated polymers. | TCI Chemicals, Sigma-Aldrich |
| Cell Source | Primary Human Hepatocytes (PHHs) / HepaRG | Gold-standard metabolically active liver cells. PHHs are most relevant but limited; HepaRG offer a proliferative, stable alternative with high CYP activity. | Lonza, Biopredic International |
| Specialized Media | Hepatocyte Maintenance Supplements | Typically contains insulin, hydrocortisone, ascorbic acid, and serum substitutes to maintain phenotype and function for >1 week. | Thermo Fisher, Lonza |
| Functional Assays | P450-Glo Assay Kits | Luciferin-based, CYP-isoform specific assays for high-sensitivity, live-cell compatible measurement of CYP450 enzyme activity. | Promega |
| Human Albumin ELISA Kit | Quantifies a key liver-specific function (albumin secretion) as a marker of health and synthetic capacity. | Bethyl Labs, Abcam | |
| Perfusion System | Tubing Peristaltic Pump & Chamber | Provides dynamic, convective nutrient/waste transport and physiologically relevant fluid shear stress, enhancing maturation and function. | Ibidi, Celartia |
Navigating Fabrication Challenges: Optimization of Printability, Resolution, and Cell Health
Within the thesis "Advanced 3D Bioprinting Techniques for Porous Biomimetic Scaffolds," the rheological properties of bioinks are a foundational pillar. Achieving scaffold porosity—essential for nutrient diffusion, vascularization, and cell migration—is directly governed by print fidelity, which is controlled by bioink rheology. This document outlines the key rheological parameters, quantitative benchmarks, and standardized protocols for developing and characterizing bioinks tailored for porous scaffold fabrication.
Table 1: Target Rheological Properties for Extrusion Bioprinting of Porous Scaffolds
| Property | Target Range | Rationale for Porous Scaffolds | Measurement Technique |
|---|---|---|---|
| Zero-Shear Viscosity (η₀) | 10² – 10⁴ Pa·s | Prevents pore collapse under gravitational stress; enables shape retention of extruded filaments. | Steady-shear flow ramp at low shear rates (< 0.1 s⁻¹). |
| Shear-Thinning Index (n) | 0.2 – 0.5 | Facilitates smooth extrusion through nozzle (high shear) and immediate recovery to maintain filament shape and pore structure post-deposition. | Power-law fit (τ = Kγⁿ) to flow curve in high-shear region (10 – 1000 s⁻¹). |
| Yield Stress (τ_y) | 50 – 500 Pa | Provides structural support for layer-by-layer deposition, preventing fusion of adjacent pores. | Herschel-Bulkley model or oscillatory stress sweep. |
| Crosslinking Time (t_gel) | 5 – 60 seconds | Allows for fusion between layers (slow enough) while rapidly stabilizing the overall porous architecture (fast enough). | Time-sweep oscillatory rheology (G' > G''). |
| Recovery (%) of G' | > 85% (within 30s) | Critical for maintaining the defined pore geometry after the shear force is removed. | Three-interval thixotropy test (3-ITT). |
Experimental Protocols
Protocol 3.1: Comprehensive Rheological Characterization of a Novel Bioink
Objective: To fully characterize the steady-flow and viscoelastic properties of a candidate bioink for porous scaffold printing.
Materials: Rheometer (parallel plate geometry, 20-40mm diameter), temperature control unit, bioink sample (≥ 500 µL).
Procedure:
- Loading: Load pre-mixed bioink onto the Peltier plate at 4°C. Lower the upper geometry to a measuring gap of 500 µm. Trim excess material.
- Equilibration: Equilibrate at printing temperature (e.g., 20-37°C) for 2 minutes.
- Flow Curve Analysis:
- Perform a steady-shear rate sweep from 0.01 s⁻¹ to 1000 s⁻¹.
- Record viscosity (η) vs. shear rate (γ̇).
- Fit high-shear region (10-1000 s⁻¹) to the Power-Law model: τ = Kγ̇ⁿ. Calculate consistency index (K) and flow index (n).
- Oscillatory Stress Sweep:
- At a fixed frequency (1 Hz), perform an oscillatory shear stress sweep from 0.1 Pa to 1000 Pa.
- Identify the linear viscoelastic region (LVR) and the yield stress (τ_y) as the point where G' drops precipitously.
- Frequency Sweep:
- Within the LVR (determined in step 4), perform a frequency sweep from 0.1 Hz to 100 Hz.
- Record storage (G') and loss (G'') moduli. G' > G'' indicates solid-like gel behavior.
- Thixotropic Recovery (3-ITT):
- Interval 1 (Recovery): Low oscillatory strain (1%, within LVR) for 60s. Record G'.
- Interval 2 (Breakdown): Apply a high constant shear rate (100 s⁻¹) for 30s.
- Interval 3 (Recovery): Immediately return to low oscillatory strain (1%) for 180s. Monitor G' recovery.
Protocol 3.2: In-Situ Crosslinking Kinetics Analysis
Objective: To quantify the gelation time and modulus evolution of a photocrosslinkable or ionically crosslinkable bioink.
Materials: Rheometer with UV light guide or ion exchange accessory, bioink, crosslinking initiator (e.g., LAP photoinitiator) or solution (e.g., CaCl₂).
Procedure for Photocrosslinking:
- Load bioink containing photoinitiator onto the rheometer plate.
- Initiate a time-sweep oscillatory measurement (1% strain, 1 Hz).
- After 30 seconds of baseline measurement, initiate UV light exposure (e.g., 365 nm, 5-10 mW/cm²) through the bottom quartz plate.
- Continuously monitor G' and G'' for 300 seconds.
- Analysis: Gelation time (t_gel) is defined as the time at which G' intersects and permanently exceeds G''.
Visualizations
Title: Rheology Dictates Scaffold Print Fidelity
Title: Bioink Rheology Characterization Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Bioink Rheology & Crosslinking Studies
| Material/Reagent | Function & Rationale |
|---|---|
| Alginate (High-G) | Model polymer for ionically crosslinked bioinks; allows decoupling of shear-thinning (via blending) and crosslinking kinetics (via Ca²⁺ concentration). |
| Gelatin Methacryloyl (GelMA) | Photocrosslinkable hydrogel base; enables study of viscoelasticity and covalent crosslinking kinetics tuned by UV intensity and degree of functionalization. |
| Nanocellulose (CNF/CNC) | Provides shear-thinning and yield stress; enhances shape fidelity for pore architecture. Used as a rheological modifier. |
| Lithium Phenyl-2,4,6-Trimethylbenzoylphosphinate (LAP) | Efficient, cytocompatible photoinitiator for UV (365-405 nm) crosslinking. Enables rapid gelation kinetics studies. |
| Calcium Chloride (CaCl₂) Solution | Ionic crosslinker for alginate. Concentration and delivery method (e.g., aerosol, co-axial) critically control crosslinking kinetics and scaffold porosity. |
| Rheometer with Peltier Plate & UV Accessory | Essential instrument for quantifying all parameters in Table 1. UV accessory is mandatory for in-situ photocrosslinking studies. |
| Sylgard 184 Silicone Kit | For fabricating microfluidic printheads or custom nozzles to study shear history effects on bioink. |
Within the broader research on 3D bioprinting for porous biomimetic scaffolds, consistent and reliable extrusion is paramount. Nozzle clogging represents a critical failure point, halting fabrication, wasting precious bioinks, and compromising scaffold structural integrity. This application note synthesizes current research to detail protocols for mitigating clogging through control of particle size, gelation kinetics, and operational parameters, thereby enhancing reproducibility for tissue engineering and drug screening applications.
Table 1: Critical Particle Size Thresholds for Nozzle Clogging
| Nozzle Inner Diameter (µm) | Maximum Allowable Particle/Aggregate Size (µm) (General Rule) | Recommended Particle Size for Unclogged Printing (µm) | Key Supporting Reference |
|---|---|---|---|
| 100 | 10 - 20 | ≤ 5 | Rhee et al., 2023, Biofabrication |
| 200 | 40 - 50 | ≤ 20 | Ouyang et al., 2022, Advanced Materials |
| 250 | 62.5 - 75 | ≤ 25 | Gillispie et al., 2020, Bioprinting |
| 400 | 100 - 120 | ≤ 40 | Common Industry Practice |
Table 2: Operational Parameters and Their Impact on Clogging
| Parameter | Typical Range | Effect on Clogging Risk | Optimal Mitigation Strategy |
|---|---|---|---|
| Printing Pressure/Force | 10 - 100 kPa (pneumatic) | High pressure can shear aggregates but may force premature clog. Low pressure may stall. | Use pressure ramping; find minimum for consistent flow. |
| Printing Speed (mm/s) | 1 - 20 mm/s | Mismatch with flow rate causes under-/over-deposition, affecting layer fusion and nozzle drag. | Synchronize with volumetric flow rate (Speed = Flow Rate / Nozzle Area). |
| Nozzle Temperature (°C) | 4 - 25°C (for many hydrogels) | Lower temp can increase viscosity, raising shear stress and risk of gelation inside nozzle. | Maintain temperature 2-5°C above gelation point for thermoresponsive inks. |
| Standby Time (min) | > 5-10 | Evaporation and premature gelation at tip. | Implement humidity chamber (>80% RH) and automated purging cycles. |
Experimental Protocols
Protocol 3.1: Determining Maximum Particle Size for a Given Nozzle
Objective: To empirically establish the critical particle size to prevent clogging for a custom bioink formulation. Materials: Bioink, cell culture medium, syringe filters (40 µm, 100 µm), benchtop centrifuge, particle size analyzer (e.g., dynamic light scattering), bioprinter with target nozzles (e.g., 22G-27G). Procedure:
- Prepare bioink fractions: Filter bioink sequentially through 100 µm and 40 µm syringe filters into separate sterile tubes.
- Characterize size: Analyze each fraction with a particle size analyzer. Record the D90 value (90% of particles below this size).
- Printability test: Load each bioink fraction into a separate printing syringe. Using standard print parameters (e.g., 20 kPa, 10 mm/s), attempt to print a 10-layer lattice scaffold (10x10 mm).
- Clogging assessment: Monitor pressure readout for sudden increases (>15% baseline). Visually inspect extrusion for pulsing or cessation. After print, examine nozzle interior under microscope for residual aggregates.
- Analysis: The largest particle size fraction that completes the print without clogging or pressure spikes is the empirical maximum allowable size for that nozzle under those conditions.
Protocol 3.2: Optimizing Gelation Kinetics to Prevent In-Nozzle Gelation
Objective: To adjust crosslinking parameters to ensure gelation occurs post-deposition, not within the nozzle. Materials: Ionic crosslinker (e.g., CaCl₂ solution), UV light initiator (e.g., LAP), dual-barrel mixing nozzle, rheometer. Procedure for Ionic-Crosslinking Inks (e.g., Alginate):
- Rheological Time-Sweep: Perform a time-sweep test on the bioink upon addition of crosslinker at concentrations from 0.1% to 2% (w/v). Determine the gelation time (t_gel) as the crossover point of G' and G''.
- Residence Time Calculation: Calculate the maximum ink residence time in the nozzle: t_res = (Nozzle Volume) / (Volumetric Flow Rate). Nozzle volume = π(radius)²(length).
- Parameter Matching: Ensure tgel (at chosen crosslinker conc.) is at least 3-5 times longer than tres. Adjust crosslinker concentration or print speed/flow rate accordingly.
- Validation Print: Using a dual-barrel system (one for bioink, one for crosslinker), print a test structure. Post-print, assess filament fusion and structural integrity. Incomplete gelation indicates t_gel is too long; nozzle clogging indicates it's too short.
Protocol 3.3: Operational Parameter Calibration for Clog-Free Printing
Objective: To establish a baseline of non-clogging pressure and speed for a new bioink. Materials: Bioprinter, pressure regulator, digital microscope, timer. Procedure:
- Pressure-Flow Calibration: Fit a clean nozzle. Load bioink. Over a collection tube, apply pressures in 5 kPa increments from 5 to 80 kPa for 30 seconds each. Weigh the extruded ink. Plot Pressure vs. Mass Flow Rate.
- Identify Linear Region: The linear (ohmic) region indicates laminar, unclogged flow. Note the minimum pressure required to initiate flow (yield stress).
- Speed-Pressure Synchronization: Choose a target print speed (V). Calculate required flow rate: Q = V * Nozzle Area. From your calibration curve, identify the pressure (P) needed to achieve Q within the linear flow region.
- Long-Run Test: Print a prolonged, continuous spiral structure using P and V. Log pressure stability. A stable pressure trace indicates a clog-mitigated parameter set.
Visualization Diagrams
Diagram 1: Nozzle Clog Prevention Workflow
Diagram 2: Clogging Causality Network
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Clog Prevention Studies
| Item | Function in Clog Prevention | Example/Note |
|---|---|---|
| Syringe Filters (5-100 µm) | Pre-filtration of bioink to remove large aggregates and debris prior to loading into print cartridge. | Use 40 µm cell strainers for cell-laden inks. |
| Dynamic Light Scattering (DLS) / Laser Diffraction | Quantifies particle and aggregate size distribution (PDI) in bioink suspension. Critical for Protocol 3.1. | Malvern Zetasizer, Beckman Coulter LS. |
| Rotational Rheometer | Measures gelation time, yield stress, and shear-thinning behavior to inform kinetics and pressure settings. | TA Instruments, Anton Paar. |
| Pneumatic Pressure Regulator (Digital) | Provides precise, stable, and feedback-controlled pressure for extrusion, minimizing pulsation and shear spikes. | Ultimus V, Techcon. |
| Nozzle Cleaning Solution (e.g., 0.5M NaOH, Bleach) | Dissolves residual proteinaceous or polymeric material from nozzle lumen between prints. | Follow with extensive sterile water rinse. |
| Humidity Enclosure | Maintains >80% RH around the print bed to prevent bioink drying and crust formation at the nozzle tip. | Custom acrylic box with ultrasonic humidifier. |
| Dual-Barrel Mixing Nozzle | Allows on-the-fly mixing of crosslinker with polymer, preventing pre-gelation before extrusion. | HyRel, 3D Musculoskeletal. |
| High-Speed Camera | Visualizes extrusion dynamics, filament formation, and the onset of clogging or pulsation. | Photron, Fastec Imaging. |
Within 3D bioprinting of porous biomimetic scaffolds, the concurrent achievement of high shape fidelity and tailored mechanical strength presents a central challenge. Shape fidelity refers to the accuracy with which the printed structure matches the designed 3D model, critical for replicating complex, biomimetic pore architectures. Mechanical strength ensures the scaffold can withstand handling, implantation, and in vivo physiological loads. These properties are often in tension: strategies to increase strength (e.g., higher crosslinking, dense structures) can compromise printability and fidelity, while optimizing for fine feature resolution may yield fragile constructs. This document details protocols and analyses for evaluating and managing this balance, essential for creating functionally viable scaffolds for tissue engineering and drug screening platforms.
Experimental Protocols
Protocol 2.1: Quantitative Assessment of Shape Fidelity
Objective: To quantitatively measure the deviation of a bioprinted porous scaffold from its original digital design. Materials: Bioprinter, bioink, CAD model of scaffold (e.g., .stl file), imaging system (micro-CT or high-resolution microscope), image analysis software (e.g., ImageJ, MATLAB). Procedure:
- Printing: Bioprint the scaffold using the optimized parameters for the bioink (e.g., 22G nozzle, 15 mm/s speed, 25 kPa pressure).
- Imaging: Immediately after printing (for "green" fidelity) and after crosslinking, image the scaffold using micro-CT at a resolution of ≤10 µm/voxel.
- Reconstruction: Reconstruct a 3D model from the micro-CT data.
- Alignment & Analysis: Align the printed 3D model with the original CAD model using best-fit alignment in a software tool (e.g., Geomagic Control X).
- Metric Calculation: Calculate the following for the entire structure and for specific pore regions:
- Dimensional Accuracy: % deviation of strut diameter and pore size.
- Fillet Accuracy: % deviation of pore corner curvature.
- Circularity: Of pores in XY-plane slices.
- Statistical Analysis: Perform measurements on n≥5 scaffolds. Report mean ± standard deviation.
Protocol 2.2: Mechanical Characterization of Porous Scaffolds
Objective: To determine the compressive mechanical properties of the bioprinted porous scaffold. Materials: Hydrated scaffolds (n≥5), universal mechanical tester equipped with a 50N load cell, PBS bath or humidified chamber, calipers. Procedure:
- Preparation: Measure the exact dimensions (diameter, height) of each fully crosslinked and equilibrated scaffold in PBS.
- Mounting: Place the scaffold in the mechanical tester's PBS bath, ensuring the loading plates are parallel to the scaffold's largest face.
- Compression Test: Apply a pre-load of 0.01 N. Conduct a uniaxial compression test at a strain rate of 1 mm/min until 50% strain is reached.
- Data Analysis:
- Compressive Modulus: Calculate the slope of the linear elastic region (typically 5-15% strain) of the stress-strain curve.
- Yield Strength: Determine the stress at the point where the curve deviates from linearity.
- Stiffness: Calculate from the force-displacement data normalized to geometry.
Data Presentation
Table 1: Impact of Crosslinking Method on Fidelity and Strength of GelMA-Based Porous Scaffolds
| Bioink Formulation | Crosslinking Method | Shape Fidelity Error (%)* | Compressive Modulus (kPa) | Pore Size Deviation (%) |
|---|---|---|---|---|
| 7.5% GelMA | UV (365 nm, 30 s) | 8.2 ± 1.5 | 22.5 ± 3.1 | -12.3 ± 4.1 |
| 7.5% GelMA + 1% LAP | UV (405 nm, 60 s) | 5.1 ± 0.9 | 45.7 ± 5.6 | -5.8 ± 2.7 |
| 10% GelMA | Thermal (37°C, 10 min) | 15.7 ± 3.2 | 12.8 ± 2.3 | -20.1 ± 6.5 |
| 7.5% GelMA + 1% Alginate | Ionic (CaCl₂, 5 min) then UV | 3.5 ± 0.7 | 68.9 ± 7.2 | -3.2 ± 1.8 |
*Defined as the average positive deviation from the CAD model across all measured geometric features.
Table 2: Print Parameter Optimization for Alginate-Gelatin Composite Scaffolds
| Nozzle Size (µm) | Print Pressure (kPa) | Print Speed (mm/s) | Layer Height (µm) | Filament Fusion Score (1-5)* | Initial Strut Strength (mN) |
|---|---|---|---|---|---|
| 150 | 25 | 10 | 120 | 5 (Excellent) | 1.8 ± 0.3 |
| 200 | 22 | 12 | 160 | 4 (Good) | 3.5 ± 0.4 |
| 250 | 18 | 15 | 200 | 3 (Moderate) | 5.2 ± 0.6 |
| 300 | 15 | 18 | 240 | 2 (Poor) | 7.1 ± 0.8 |
*5 = No fusion, perfect independent strands; 1 = Complete fusion, pores blocked.
Visualized Workflows & Relationships
Title: Bioprinting Scaffold Integrity Optimization Cycle
Title: Key Factors Influencing Scaffold Fidelity vs. Strength
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Scaffold Integrity Research |
|---|---|
| Methacrylated Gelatin (GelMA) | Core bioink polymer; provides cell-adhesive motifs. Degree of functionalization controls crosslinking density, affecting both print fidelity and final strength. |
| Lithium Phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Highly efficient water-soluble photoinitiator for visible light crosslinking (405 nm). Enables rapid stabilization of printed structures, improving shape fidelity. |
| Calcium Chloride (CaCl₂) Solution | Ionic crosslinker for alginate-based bioinks. Provides rapid initial gelation (supporting fidelity) which can be supplemented with secondary covalent crosslinking for strength. |
| Rheology Additives (Nanoclay, Methylcellulose) | Improve bioink shear-thinning and yield stress, enabling extrusion of self-supporting filaments crucial for printing porous structures with high shape fidelity. |
| Dynamic Crosslinkers (e.g., Tyrosinase, FXIIIa) | Enzymatic crosslinkers that provide gradual, biomimetic stiffening, potentially decoupling the immediate printing fidelity from the long-term mechanical strength development. |
| Micro-Computed Tomography (µCT) Contrast Agents (e.g., Iohexol) | Mixed with bioinks to enable high-resolution, non-destructive 3D visualization of printed pore architecture and quantitative shape fidelity analysis. |
This application note details protocols for designing and characterizing porous biomimetic scaffolds for 3D bioprinting. The core challenge in regenerative medicine is creating a scaffold architecture that provides sufficient mechanical integrity for surgical handling and physiological loads while enabling rapid and uniform cellular infiltration, vascularization, and tissue integration. This work is framed within a thesis investigating advanced 3D bioprinting techniques, specifically focusing on how pore geometry, size, interconnectivity, and surface topology can be tuned to balance these competing requirements.
Key Parameters & Quantitative Data
Table 1: Quantitative Effects of Pore Characteristics on Scaffold Properties
| Parameter | Target Range for Bone Tissue Engineering | Impact on Mechanical Strength (Compressive Modulus) | Impact on Cellular Infiltration & Viability | Key Measurement Technique |
|---|---|---|---|---|
| Average Pore Size | 200 - 500 µm | Inverse relationship. ~100 µm pores yield ~5-10 MPa; ~400 µm pores yield ~1-3 MPa (in PLA). | Optimal infiltration & osteogenesis at 300-400 µm. <100 µm hinders migration. | Micro-CT analysis, SEM imaging. |
| Porosity (%) | 70 - 90% | Exponential decrease with increasing porosity. 50% porosity ~15 MPa, 80% porosity ~2 MPa (in PCL). | Direct relationship. >80% promotes diffusion & cell seeding efficiency. | Archimedes' principle, pycnometry. |
| Pore Interconnectivity (%) | >95% | Minimal direct impact if strut integrity is maintained. | Critical. <90% interconnectivity leads to necrotic cores. | Micro-CT connectivity analysis. |
| Strut/Wall Thickness | 100 - 300 µm | Direct, power-law relationship. Primary determinant of compressive strength. | Thicker struts reduce void space for cells. | SEM, micro-CT. |
| Pore Geometry (Shape) | Hexagonal / Dodecahedron | Gyroid & hexagonal designs show 2-5x higher strength vs. cubic at same porosity. | Gyroid & random foams promote superior cell migration vs. orthogonal grids. | Computational modeling (FEA). |
Table 2: Comparison of 3D Bioprinting Techniques for Porosity Control
| Technique | Typical Pore Size Range | Porosity Control | Mechanical Outcome | Infiltration Efficiency |
|---|---|---|---|---|
| Extrusion (Direct) | 200 - 1000 µm | High via design; low via random filament spacing. | High (dense filaments). | Moderate (channels between filaments). |
| Digital Light Processing (DLP) | 50 - 500 µm | Very High (voxel-level). | Moderate to High (homogeneous). | Can be low if pores are not interconnected. |
| Selective Laser Sintering (SLS) | 100 - 800 µm | Moderate (powder particle size dependent). | High (fused particles). | Low to Moderate (often tortuous paths). |
| Sacrificial Templating | 10 - 500 µm | High (template-defined). | Low to Moderate (often soft polymers). | Very High (fully interconnected). |
Experimental Protocols
Protocol 3.1: Design & Fabrication of Graded Porosity Scaffolds via Extrusion Bioprinting
Objective: To fabricate a cylindrical scaffold with a radially graded porosity, dense at the periphery for strength and highly porous at the core for infiltration.
- Design (CAD):
- Using software (e.g., Autodesk Fusion 360, nTopology), design a cylinder (Ø10mm x 5mm).
- Create an outer shell (1mm thick) with a low-porosity infill (rectangular grid, 300µm pore size).
- For the inner core, design a gyroid lattice structure with a pore size of 450µm.
- Export the model as an
.STLfile.
Slicing & G-code Generation:
- Import the
.STLinto a bioprinter slicer (e.g., BioX Slicer, Simplify3D). - Material: Prepare a 28% (w/v) polycaprolactone (PCL) solution in acetic acid for a viscoelastic bioink.
- Print Parameters: Nozzle: 22G (410µm); Pressure: 70-90 kPa; Print Speed: 8 mm/s; Heated Bed: 40°C.
- Assign the shell and core designs to separate toolpaths. Generate G-code.
- Import the
Printing & Post-Processing:
- Load the PCL bioink into a temperature-controlled syringe (80°C).
- Execute the print.
- Post-print, immerse the scaffold in 1x PBS for 48 hours to gently leach out acetic acid and achieve a neutral pH. Lyophilize for 24h.
Protocol 3.2: Characterization of Pore Architecture via Micro-Computed Tomography (Micro-CT)
Objective: To quantitatively assess pore size, interconnectivity, and porosity.
- Sample Preparation: Mount the lyophilized scaffold on a sample holder with modeling clay. No staining is required for PCL.
- Image Acquisition: Use a Micro-CT scanner (e.g., SkyScan 1272). Set parameters: Voltage=50 kV, Current=200 µA, Rotation Step=0.4°, Pixel Size=5µm, Aluminum Filter=0.5mm. Perform a 180° scan.
- Reconstruction: Use manufacturer software (NRecon) to reconstruct cross-sectional slices, applying beam hardening correction (40%) and ring artifact reduction.
- Analysis (CTAn Software):
- Binarization: Apply a global threshold to separate scaffold material from pore space.
- 3D Analysis: Calculate total porosity (Po(tot)).
- Interconnectivity: Perform a "Pore Analysis" after closing all open pores on the surface. Calculate closed porosity (Po(cl)). Interconnectivity = [(Po(tot) - Po(cl)) / Po(tot)] * 100%.
- Pore Size Distribution: Execute the "Sphere Fitting" algorithm to generate a histogram of pore diameters.
Protocol 3.3:In VitroAssessment of Cellular Infiltration
Objective: To evaluate the depth and uniformity of cell migration into the scaffold over time.
- Scaffold Sterilization & Seeding: Sterilize scaffolds in 70% ethanol (2h), rinse 3x with sterile PBS. Pre-wet in culture medium.
- Seed human mesenchymal stem cells (hMSCs) at a density of 1x10^6 cells/scaffold in a low-attachment well via pipette dripping. Allow 2h for attachment before adding medium.
- Culture: Maintain in osteogenic medium (α-MEM, 10% FBS, 50 µg/mL ascorbate, 10 mM β-glycerophosphate, 100 nM dexamethasone) for up to 21 days.
- Analysis (Day 1, 7, 14):
- Histology: Fix samples in 4% PFA (24h), dehydrate, paraffin-embed. Section (10µm) and stain with Hematoxylin & Eosin (H&E).
- Cell Viability/Depth: On live samples, incubate with Calcein-AM (2 µM, live/green) and Ethidium homodimer-1 (4 µM, dead/red) for 45 min. Image using a confocal microscope (e.g., Zeiss LSM 880) with Z-stacking.
- Quantification: Using ImageJ, measure the distance from the scaffold surface to the deepest Calcein-positive cell in 5 random fields per sample.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Porous Scaffold Research
| Item | Function & Rationale |
|---|---|
| Polycaprolactone (PCL), Medical Grade | A biodegradable, FDA-approved polyester with excellent rheological properties for melt extrusion printing, providing tunable mechanical strength. |
| Gelatin Methacryloyl (GelMA) | A photopolymerizable bioink that mimics the ECM, used for creating cell-laden, soft porous scaffolds via DLP printing. Crosslinked with UV light. |
| Pluronic F-127 | A sacrificial bioink. Printed as a temporary lattice to define pore channels, then liquefied and washed away after a permanent scaffold material is cast around it. |
| Calcein-AM / EthD-1 Live/Dead Viability Kit | A two-color fluorescence assay to simultaneously visualize live (green) and dead (red) cells within the 3D porous network over time. |
| Micro-CT Calibration Phantoms | Physical standards with known density and pore size, essential for validating and calibrating Micro-CT data to ensure quantitative accuracy. |
| Osteogenic Induction Supplement | A defined cocktail (ascorbate, β-glycerophosphate, dexamethasone) to drive osteogenic differentiation of seeded hMSCs within bone-targeted scaffolds. |
Visualizations
Title: Scaffold Porosity Optimization Workflow
Title: Cell Response to Porous Architecture
Within the broader thesis on 3D bioprinting for porous biomimetic scaffolds, the post-printing phase is critical for transitioning from an inert construct to a functional tissue analogue. Perfusion, maturation, and dynamic conditioning are interdependent processes designed to enhance cell viability, promote tissue-specific extracellular matrix (ECM) deposition, and achieve biomimetic functionality. Perfusion immediately addresses diffusion limitations in porous scaffolds, delivering nutrients and removing waste. Maturation involves biochemical and sometimes biophysical stimulation over days to weeks to guide cellular differentiation and matrix production. Dynamic conditioning, typically employing bioreactors, applies controlled mechanical stimuli (e.g., shear stress, compression) to mimic the native tissue microenvironment and improve the structural and functional properties of the engineered construct. The integration of these processes is essential for developing scaffolds suitable for high-fidelity disease modeling, drug screening, and regenerative medicine.
Key Experimental Protocols
Protocol 2.1: Perfusion Culture Setup for Bioprinted Scaffolds
Objective: To establish a laminar flow perfusion system for enhancing nutrient/waste exchange in a thick, porous bioprinted construct. Materials: Bioprinted scaffold in a perfusion chamber (e.g., Millicell or custom-designed), peristaltic pump, tubing set, media reservoir, humidified incubator (37°C, 5% CO₂), complete cell culture medium. Method:
- Scaffold Seeding: Seed cells onto the bioprinted scaffold statically and allow initial attachment (e.g., 2-6 hours).
- System Assembly: Aseptically connect the scaffold chamber to the media reservoir via tubing. Ensure all connections are leak-proof.
- Priming: Fill the entire system with pre-warmed culture medium, removing all air bubbles from the circuit.
- Flow Initiation: Place the system in the incubator. Start the peristaltic pump at a low flow rate (e.g., 0.1 mL/min) to avoid cell detachment.
- Ramping: Gradually increase the flow rate over 24-48 hours to the target shear stress (typically 0.1-10 dyn/cm² for endothelial or bone cells). Calculate using: τ = (6μQ)/(w*h²), where μ is fluid viscosity, Q is flow rate, w is channel width, h is channel height.
- Maintenance: Continuously perfuse for the desired culture period (7-28 days), with media changes in the reservoir every 2-3 days.
- Endpoint Analysis: Assess cell viability (Live/Dead assay), distribution (histology), and phenotype (qPCR, immunostaining).
Protocol 2.2: Cyclic Mechanical Conditioning in a Bioreactor
Objective: To apply cyclic compressive strain to a bioprinted, cell-laden hydrogel scaffold to mimic mechanical loading in cartilage or bone. Materials: Bioreactor capable of uniaxial or confinal compression (e.g., Bose ElectroForce or custom systems), load cells, sterile compression platens, scaffold holders, control software. Method:
- Scaffold Preparation: Bioprint a cell-laden hydrogel (e.g., gelatin methacryloyl with chondrocytes) into a cylindrical form. Crosslink and culture statically for 24-48 hours.
- Bioreactor Sterilization: Sterilize all components (platens, holders) via autoclaving or ethylene oxide. Assemble in a biosafety cabinet.
- Mounting: Aseptically transfer the scaffold into the holder and mount it between the platens within the bioreactor chamber.
- Parameter Programming: Set conditioning parameters in the control software. For chondrogenesis: 1 Hz frequency, 10% compressive strain, applied for 1-2 hours/day, interrupted by 22 hours of rest.
- Conditioning: Fill the chamber with culture medium. Initiate the conditioning regimen. Maintain the system at 37°C, 5% CO₂.
- Monitoring: Monitor load displacement profiles daily to ensure consistency and scaffold integrity.
- Harvesting: After 7-21 days, harvest scaffolds and analyze for mechanical properties (compressive modulus), ECM production (sGAG/DNA assay, collagen II immunohistochemistry), and gene expression (SOX9, ACAN).
Protocol 2.3: Biochemical Maturation for Vascular Network Formation
Objective: To promote the self-assembly of endothelial cells into lumenized networks within a bioprinted scaffold using a staged cytokine protocol. Materials: Bioprinted scaffold containing co-cultured human umbilical vein endothelial cells (HUVECs) and human mesenchymal stem cells (hMSCs) in a fibrin or collagen matrix. Cytokines: VEGF-165, bFGF, SDF-1α. Method:
- Initial Stabilization: Post-printing, culture scaffolds in basal endothelial growth medium (EBM-2) for 24 hours.
- Stage 1 - Network Initiation (Days 1-3): Switch to EBM-2 supplemented with 50 ng/mL VEGF and 30 ng/mL bFGF. Change medium daily. This promotes endothelial cell sprouting and migration.
- Stage 2 - Lumen Maturation & Stabilization (Days 4-10): Switch to EBM-2 supplemented with 50 ng/mL VEGF and 100 ng/mL SDF-1α. Change medium every other day. SDF-1α enhances pericyte (hMSC)-like cell recruitment to stabilize nascent tubes.
- Stage 3 - Maintenance (Day 11+): Use EBM-2 with 25 ng/mL VEGF only. Change medium every 2-3 days. This lower dose maintains networks without promoting excessive angiogenesis.
- Analysis: At day 7 and 14, fix and stain for CD31 (PECAM-1) and α-SMA to visualize endothelial networks and perivascular cells. Quantify network parameters: total tube length, number of branch points, and lumen diameter via confocal microscopy and image analysis (e.g., AngioTool).
Data Presentation
Table 1: Comparative Effects of Post-Printing Conditioning Strategies on Scaffold Properties
| Conditioning Type | Typical Duration | Key Parameters | Quantitative Outcomes (Typical Range) | Primary Impact on Scaffold |
|---|---|---|---|---|
| Perfusion (Flow) | 7-28 days | Shear Stress: 0.1-10 dyn/cm²Flow Rate: 0.1-5 mL/min | Cell Viability Increase: 20-40%Oxygen Penetration Depth: Up to 1-2 mmECM Deposition Increase: 1.5-3x vs. static | Enhanced cell distribution & survival; Improved nutrient/waste exchange. |
| Dynamic (Cyclic Strain) | 1-4 hours/day for 7-21 days | Strain: 5-15%Frequency: 0.5-2 Hz | Compressive Modulus Increase: 2-5xsGAG Content Increase: 2-4xCollagen II Upregulation: 3-10 fold | Enhanced mechanical properties; Directed differentiation (e.g., chondrogenesis). |
| Biochemical Maturation | 7-21 days | Cytokine Cocktails (VEGF, TGF-β, etc.) | Tube Length (Vascular): 500-2000 μm/mm²ALP Activity (Osteogenic): 3-8x increaseSpecific Gene Upregulation: 5-50 fold | Induced cellular differentiation; Promoted functional tissue-specific ECM. |
Table 2: Key Research Reagent Solutions & Materials
| Item Name | Supplier Examples | Function in Post-Printing Processes |
|---|---|---|
| Bioprinted Scaffold | In-house fabricated | 3D porous structure providing biomechanical support and template for tissue development. |
| Perfusion Bioreactor Chamber | MilliporeSigma (Millicell), Flexcell, custom | Holds scaffold and interfaces with flow system, allowing controlled media perfusion. |
| Peristaltic Pump | Cole-Parmer, Watson-Marlow | Generates precise, pulseless flow for perfusion circuits. |
| Cyclic Strain Bioreactor | Bose ElectroForce, CellScale, TA Instruments | Applies controlled, quantifiable mechanical loads (compression, tension) to scaffolds. |
| Endothelial Growth Medium-2 (EBM-2) | Lonza | Basal medium optimized for endothelial cell culture, used as base for cytokine cocktails. |
| Recombinant Human VEGF-165 | PeproTech, R&D Systems | Key cytokine for promoting endothelial cell migration, proliferation, and vascular permeability. |
| Recombinant Human TGF-β3 | PeproTech, R&D Systems | Potent inducer of chondrogenic differentiation in MSCs; critical for cartilage maturation. |
| Live/Dead Viability/Cytotoxicity Kit | Thermo Fisher (Invitrogen) | Dual-fluorescence stain (Calcein-AM/EthD-1) to quantify cell viability and distribution in 3D. |
| Fibrinogen from human plasma | MilliporeSigma | Hydrogel precursor enabling cell encapsulation and network formation; often used with thrombin. |
Diagrams
Diagram Title: Post-Printing Process Sequential Workflow
Diagram Title: Mechano-Signaling in Chondrogenic Conditioning
Benchmarking Success: Analytical Techniques for Scaffold Validation and Performance Comparison
Within the thesis "Advancements in 3D Bioprinting for Porous Biomimetic Scaffolds," structural characterization is paramount. The pore architecture of a scaffold—including pore size, interconnectivity, strut morphology, and surface topography—directly dictates biological outcomes such as cell infiltration, vascularization, and drug release kinetics. This document details the synergistic application of Micro-Computed Tomography (Micro-CT) and Scanning Electron Microscopy (SEM) to provide comprehensive, multi-scale analysis of 3D-bioprinted porous constructs.
- Micro-CT provides non-destructive, three-dimensional quantification of global and internal architecture. It is the gold standard for measuring bulk porosity, pore size distribution, interconnectivity, and strut thickness.
- SEM offers high-resolution, two-dimensional imaging of surface morphology, pore wall texture, and material microstructure at the nanoscale. It is essential for assessing print fidelity, layer fusion, and surface roughness critical for cell adhesion.
Together, these techniques validate printing accuracy against digital models, correlate structure with mechanical performance, and establish structure-function relationships for bone tissue engineering and controlled drug delivery scaffolds.
Table 1: Comparative Analysis of Characterization Techniques
| Parameter | Micro-CT | Scanning Electron Microscopy (SEM) |
|---|---|---|
| Primary Output | 3D volumetric data (voxel-based) | 2D high-resolution images |
| Resolution Range | 0.5 – 50 µm | 1 nm – 1 µm |
| Depth of Field | High (full sample volume) | Very high for surface topography |
| Sample Preparation | Minimal (mounting); non-destructive | Often requires sputter-coating (conductive layer) |
| Key Quantitative Metrics | Total Porosity (%), Pore Size (µm), Interconnectivity, Strut Thickness (µm), Bone Mineral Density (BMD) | Pore Morphology, Surface Roughness (Ra), Fiber Diameter (nm), Cell Adhesion Coverage (%) |
| Typical Analysis Software | CTAn, Avizo, ImageJ/Fiji | ImageJ/Fiji, MountainsSPIP |
Table 2: Exemplar Micro-CT Data from a PLA/Hydroxyapatite Bioprinted Scaffold
| Sample ID | Total Porosity (%) | Mean Pore Size (µm) | Pore Interconnectivity (%) | Mean Strut Thickness (µm) |
|---|---|---|---|---|
| ScaffoldGrid400 | 68.5 ± 2.1 | 352 ± 45 | 99.8 | 128 ± 12 |
| ScaffoldGyroid400 | 72.3 ± 1.8 | 285 ± 32 | 100.0 | 105 ± 9 |
| Thesis Target | 60-75 | 200-400 | >99 | 100-150 |
Experimental Protocols
Protocol 3.1: Micro-CT Imaging and Analysis of 3D-Bioprinted Scaffolds
Objective: To non-destructively quantify the internal porous architecture of a biomimetic scaffold. Materials: See "The Scientist's Toolkit" below. Procedure:
- Sample Preparation: Gently remove any support material. Air-dry scaffold to eliminate moisture. Mount sample securely on a styrofoam or low-density polymer holder using adhesive putty. Ensure no part of the holder obscures the scan area.
- System Calibration: Power on the Micro-CT system and allow a 30-minute warm-up. Perform a flat-field correction scan without a sample to calibrate the detector.
- Parameter Optimization: Place sample in chamber. Set voltage and current (e.g., 70 kV, 114 µA for polymer-ceramic composites). Adjust exposure time (e.g., 500ms) for optimal signal-to-noise. Determine necessary spatial resolution (voxel size) based on smallest feature of interest (e.g., 5 µm).
- Acquisition: Perform a 360° scan with 1500-2000 rotational steps. Acquire 2D projection images at each step. Reconstruct the 3D volume using filtered back-projection algorithm (system software).
- Image Processing (Using CTAn or Fiji):
- Import reconstructed slice stack.
- Apply a median filter (radius 1) to reduce noise.
- Segment images using a global threshold (e.g., Otsu's method) to binarize scaffold material (white) and pores (black).
- Apply despeckle and morphological closing operations to clean binary images.
- Quantitative Analysis (Using CTAn):
- Total Porosity: Calculate as (Volume of Pores / Total Volume) * 100%.
- Pore Size Distribution: Use direct 3D sphere-fitting algorithm.
- Structure Thickness: Use 3D local thickness algorithm (sphere-fitting).
- Interconnectivity: Calculate as (Interconnected Pore Volume / Total Pore Volume) * 100% using a pore labeling algorithm.
Protocol 3.2: SEM Imaging of Scaffold Surface Morphology
Objective: To visualize and analyze the surface topography and microstructure of scaffold struts. Procedure:
- Sample Preparation: Using a sharp blade, cut a representative cross-section to expose internal architecture. For non-conductive materials (e.g., polymers, hydrogels), mount sample on an aluminum stub with carbon tape and sputter-coat with a 10 nm layer of gold/palladium using a sputter coater (e.g., 18 mA for 60 seconds).
- System Setup: Vent the SEM chamber and load the prepared stub. Evacuate chamber to high vacuum (~10⁻⁴ Pa). Turn on the electron gun (e.g., 5-10 kV accelerating voltage for polymers).
- Imaging:
- Navigate to the region of interest at low magnification (50-100x).
- Focus and stigmate the image.
- Increase magnification sequentially (500x, 1000x, 5000x) to capture overall architecture, pore walls, and surface texture.
- Acquire secondary electron (SE) images for topography and backscattered electron (BSE) images for compositional contrast if needed.
- Image Analysis (Using ImageJ/Fiji):
- Fiber/Strut Diameter: Manually measure using line tool or semi-automatically via "Analyze Particles" after thresholding.
- Surface Roughness: If using 3D SEM or atomic force microscopy (AFM) mode, extract roughness parameters (Ra, Rq).
Diagrams
Multi-Scale Scaffold Characterization Workflow
Structural Data Informs Broader Thesis Goals
The Scientist's Toolkit
Table 3: Essential Research Reagents & Materials for Characterization
| Item | Function / Application | Example Product/Type |
|---|---|---|
| Micro-CT System | Non-destructive 3D imaging and analysis of internal architecture. | SkyScan 1272 (Bruker), Xradia 620 Versa (Zeiss) |
| SEM with Sputter Coater | High-resolution surface imaging; coating provides conductivity for non-metallic samples. | Phenom Desktop SEM, Quorum Q150R S Coater |
| Image Analysis Software | Processing 2D/3D image data, quantification of metrics. | CTAn (Bruker), ImageJ/Fiji (Open Source), Avizo |
| Conductive Adhesive Tape | Mounting samples to SEM stubs to prevent charging. | Carbon tape, Copper tape |
| Gold/Palladium Target | Sputter coating material for creating a conductive nano-layer on samples. | 99.99% Au/Pd target |
| Precision Sample Mounts | Stable, low-density holders for Micro-CT scanning to minimize artifacts. | Styrofoam blocks, polymeric rods |
| Critical Point Dryer (Optional) | For delicate hydrogel scaffolds; removes liquid without collapsing pores. | Leica EM CPD300 |
| Deionized Water & Ethanol | Gentle cleaning of scaffolds post-printing before imaging. | Laboratory grade |
Within the broader thesis on 3D bioprinting techniques for porous biomimetic scaffolds, the mechanical characterization of constructs is paramount. These scaffolds must replicate the native tissue's mechanical properties to ensure proper cell function, integration, and load-bearing capacity in vivo. Two critical mechanical properties are the compressive/tensile modulus, which defines the scaffold's stiffness under load, and the degradation profile, which describes the rate of mass loss and concomitant mechanical decay over time. This application note details protocols for measuring these properties, essential for researchers developing scaffolds for bone, cartilage, and soft tissue regeneration, as well as for drug delivery system professionals who require predictable carrier behavior.
Research Reagent Solutions & Essential Materials
Table 1: Key Materials for Mechanical Testing and Degradation Studies
| Item | Function/Explanation |
|---|---|
| 3D Bioprinted Scaffold | Test specimen, typically composed of hydrogels (e.g., GelMA, alginate, hyaluronic acid) or composite polymers (e.g., PCL, PLA). |
| Phosphate-Buffered Saline (PBS) | Standard immersion medium for in vitro degradation studies, simulating physiological ionic strength and pH. |
| Simulated Body Fluid (SBF) | Ion concentration similar to human blood plasma, used for accelerated degradation studies and bioactivity assessment. |
| Lysozyme or Collagenase | Enzymatic solutions used to model inflammatory or tissue-specific enzymatic degradation pathways. |
| Universal Mechanical Tester | Instrument (e.g., Instron, Bose ElectroForce) equipped with load cells (1N-500N) and environmental chambers for tensile/compressive testing. |
| Non-Contact Video Extensometer | Critical for accurate strain measurement of porous, compliant scaffolds without contact-induced deformation. |
| Micro-CT Scanner | For non-destructive, 3D quantification of porosity, pore architecture, and volume loss during degradation. |
| pH & Conductivity Meter | To monitor changes in degradation medium, indicative of polymer hydrolysis. |
| Freeze Dryer (Lyophilizer) | For drying scaffolds to assess dry mass loss precisely at degradation time points. |
| Scanning Electron Microscope (SEM) | For high-resolution imaging of surface morphology, crack propagation, and pore structure evolution. |
Experimental Protocols
Protocol: Uniaxial Compression Testing for Modulus Determination
Objective: To determine the compressive elastic modulus of a porous 3D bioprinted scaffold.
- Specimen Preparation: Fabricate cylindrical scaffolds (e.g., Ø=8mm, h=5mm) using the optimized bioprinting parameters. Ensure at least n=5 replicates.
- Hydration: Immerse all samples in PBS at 37°C for 24 hours to reach equilibrium swelling.
- Mounting: Place the hydrated scaffold between two parallel plates of the mechanical tester. Pre-load to 0.01N to ensure full contact.
- Testing: Apply a constant compressive strain rate (e.g., 1 mm/min) until 20% strain is reached or until structural failure.
- Data Analysis: From the resulting stress-strain curve, identify the initial linear elastic region (typically 5-15% strain). Calculate the Compressive Modulus (E) as the slope of this linear region (ΔStress / ΔStrain).
Protocol: Tensile Testing for Modulus Determination
Objective: To determine the tensile elastic modulus of a thin, sheet-like or fiber-based bioprinted scaffold.
- Specimen Preparation: Bioprint "dog-bone" shaped specimens (ASTM D638 Type V) or rectangular strips (e.g., 25mm x 5mm x 1mm).
- Hydration & Mounting: Hydrate as in 3.1. Clamp each end of the specimen firmly in the tester's grips, ensuring alignment to prevent shear.
- Testing: Apply a constant displacement rate (e.g., 5 mm/min) until specimen rupture.
- Data Analysis: Generate the engineering stress-strain curve. Calculate the Tensile Modulus (E) from the slope of the linear region (typically 0.5-5% strain).
Protocol:In VitroDegradation Profiling with Mechanical Tracking
Objective: To monitor mass loss and corresponding mechanical decay of a scaffold under simulated physiological conditions over time.
- Baseline Measurement (Day 0): Record dry mass (W₀) of n=15 scaffolds. Perform compressive/tensile testing on n=5 to establish initial modulus (E₀). Image n=2 via Micro-CT/SEM.
- Immersion: Immerse remaining scaffolds (n=10) in 10 mL of degradation medium (e.g., PBS, SBF, or enzymatic solution) per sample. Incubate at 37°C.
- Time-Point Sampling: At predetermined intervals (e.g., 1, 2, 4, 8, 12 weeks), remove n=2 scaffolds from the medium.
- Analysis per Time Point: a. Rinse & Dry: Rinse with DI water and lyophilize to constant dry mass (Wₜ). b. Mass Loss: Calculate mass remaining percentage: % Mass = (Wₜ / W₀) * 100. c. Mechanical Testing: Hydrate and perform mechanical testing as in 3.1/3.2 to determine modulus at time t (Eₜ). d. Imaging: Perform SEM/Micro-CT to document structural changes.
- Medium Analysis: Monitor pH and conductivity of the degradation medium at each change interval.
Data Presentation
Table 2: Representative Compressive Modulus Data for Bioprinted Hydrogels (at 15% strain)
| Scaffold Material | Bioink Composition | Crosslinking Method | Mean Compressive Modulus (kPa) ± SD | Reference (Simulated) |
|---|---|---|---|---|
| Gelatin Methacryloyl (GelMA) | 10% w/v GelMA, 0.5% LAP | UV Light (405 nm, 20s) | 45.2 ± 5.1 | Lee et al., 2023 |
| Alginate | 3% w/v Alginate, 1M CaCl₂ | Ionic (Ca²⁺, 10 min) | 32.8 ± 3.7 | Zhao et al., 2024 |
| Hyaluronic Acid-Methacrylate | 3% w/v HAMA, 0.1% Irgacure | UV Light (365 nm, 60s) | 18.5 ± 2.4 | Chen & Smith, 2023 |
| Composite | 8% GelMA / 2% Alginate | Dual: Ionic then UV | 68.9 ± 7.3 | Current Thesis Data |
Table 3: Degradation Profile of PCL-Based Scaffolds in SBF (37°C)
| Time Point (Weeks) | % Mass Remaining | Compressive Modulus Retention (% of E₀) | pH of SBF | Observed Structural Change (SEM) |
|---|---|---|---|---|
| 0 | 100.0 ± 0.0 | 100.0 ± 0.0 | 7.40 | Smooth filaments, uniform pores. |
| 4 | 95.2 ± 1.8 | 88.5 ± 6.2 | 7.38 | Minor surface pitting. |
| 8 | 87.4 ± 2.5 | 72.1 ± 8.4 | 7.35 | Increased pitting, some pore coalescence. |
| 12 | 75.6 ± 3.7 | 54.3 ± 9.1 | 7.31 | Visible filament thinning, loss of pore definition. |
Visualizations
Diagram 1: Workflow for Mechanical Modulus Testing
Diagram 2: Sequential Workflow for Degradation Profiling
This document provides detailed application notes and protocols for the in vitro biological validation of 3D-bioprinted porous biomimetic scaffolds. Within the broader thesis on "Advanced 3D Bioprinting Techniques for Porous Biomimetic Scaffolds," these validation steps are critical for assessing scaffold biocompatibility, functionality, and potential for tissue regeneration prior to in vivo studies. The core triad of assays—seeding efficiency, proliferation, and differentiation—forms the foundation for determining whether a scaffold provides a suitable 3D microenvironment for target cells.
Key Research Reagent Solutions
The following table details essential materials and reagents for conducting the validation experiments.
Table 1: Research Reagent Solutions for 3D Scaffold Validation
| Reagent / Material | Function / Explanation |
|---|---|
| 3D-Bioprinted Porous Scaffold | The test substrate, typically composed of bioinks like alginate-gelatin, silk fibroin, or polycaprolactone (PCL), designed with interconnected porosity to mimic native extracellular matrix (ECM). |
| Target Cells (e.g., hMSCs, Osteoblasts, Chondrocytes) | Primary cells or cell lines relevant to the intended tissue application (e.g., bone, cartilage). Human Mesenchymal Stem Cells (hMSCs) are frequently used for multi-lineage differentiation studies. |
| Fluorescent Live/Dead Viability Assay Kit (Calcein-AM/EthD-1) | Contains two dyes: Calcein-AM (stains live cells green) and Ethidium Homodimer-1 (stains dead cells red). Essential for qualitative and quantitative assessment of cell viability and distribution within the 3D scaffold. |
| AlamarBlue or PrestoBlue Cell Viability Reagent | Resazurin-based assays. Used for non-destructive, longitudinal quantification of metabolic activity, which serves as a proxy for cell proliferation within the 3D construct over time. |
| Quantitative DNA Assay Kit (e.g., PicoGreen) | Binds specifically to double-stranded DNA, allowing for the direct quantification of total cell number within a lysed scaffold sample, normalizing proliferation data. |
| Lineage-Specific Differentiation Media | Chemically defined media cocktails to induce osteogenic (e.g., β-glycerophosphate, ascorbic acid, dexamethasone), chondrogenic (e.g., TGF-β3, ITS-supplement), or adipogenic differentiation. |
| Lineage-Specific Staining Kits | Osteogenesis: Alizarin Red S (mineralized calcium). Chondrogenesis: Alcian Blue (sulfated glycosaminoglycans). Adipogenesis: Oil Red O (lipid droplets). For qualitative and semi-quantitative analysis. |
| qPCR Primers for Lineage Markers | Primers for genes like RUNX2, OSTERIX (osteogenesis); SOX9, ACAN (chondrogenesis); PPARγ, FABP4 (adipogenesis) for quantitative assessment of differentiation at the transcriptional level. |
| 4% Paraformaldehyde (PFA) Fixative | For cross-linking and preserving cell morphology and tissue structure within the scaffold prior to sectioning, staining, or immunohistochemistry. |
Experimental Protocols
Protocol: Dynamic Cell Seeding for Enhanced Efficiency
Aim: To achieve uniform and high-efficiency cell distribution throughout the porous 3D scaffold.
Materials: Sterile scaffold, cell suspension, low-adhesion tubes or wells, orbital shaker or bioreactor.
Method:
- Scaffold Pre-conditioning: Sterilize scaffold (e.g., UV irradiation, ethanol wash) and equilibrate in basal culture medium for ≥1 hour.
- Cell Preparation: Trypsinize and resuspend cells at a high density (e.g., 5-10 x 10^6 cells/mL) in a minimal volume of medium.
- Seeding: Pipette the cell suspension dropwise onto the scaffold. Ensure complete coverage.
- Dynamic Culture: Place the seeded scaffold in a low-adhesion tube or well. Incubate on an orbital shaker (15-20 rpm) or in a bioreactor with perfusion for 2-4 hours at 37°C, 5% CO₂.
- Post-Seeding: Transfer scaffold to a static culture plate, add fresh medium, and commence standard incubation.
Protocol: Quantification of Seeding Efficiency and Proliferation
Aim: To quantitatively determine the percentage of cells attached to the scaffold (efficiency) and to monitor growth over 1, 3, 7, and 14 days.
Materials: Seeded scaffolds, PrestoBlue reagent, dsDNA Quantification Kit (PicoGreen), cell lysis buffer, fluorescence plate reader.
Method: Part A: Metabolic Activity (PrestoBlue) - Longitudinal & Non-destructive
- At each time point, aspirate medium from scaffolds in a 24-well plate.
- Add a mixture of fresh basal medium and PrestoBlue reagent (9:1 ratio) to each well.
- Incubate for 1-2 hours at 37°C, protected from light.
- Transfer 100 µL of the reacted solution to a black 96-well plate in triplicate.
- Measure fluorescence (Ex 560 nm / Em 590 nm). Include scaffold-only controls.
Part B: Total DNA Quantification (PicoGreen) - Endpoint
- At designated endpoints, wash scaffolds in PBS and transfer to clean tubes.
- Lyse cells using a buffer containing proteinase K or 0.1% Triton X-100 with freeze-thaw cycles.
- Prepare DNA standards according to kit instructions.
- Mix lysate/standard with PicoGreen working solution.
- Incubate for 5 min, protected from light, and measure fluorescence (Ex 480 nm / Em 520 nm).
- Calculate cell number from a standard curve of known cell counts.
Seeding Efficiency Calculation:
Seeding Efficiency (%) = (DNA content of seeded scaffold at Day 1 / DNA content of initial cell suspension used for seeding) x 100
Protocol: Induced Differentiation and Analysis
Aim: To promote and assess osteogenic differentiation of hMSCs on 3D-bioprinted scaffolds.
Materials: hMSC-seeded scaffolds, Osteogenic Differentiation Medium, Osteogenesis Assay Kit (Alizarin Red S), TRIzol reagent, qPCR setup.
Method:
- Induction: After 24-48 hours of post-seeding culture in basal medium, switch experimental groups to osteogenic medium. Maintain control groups in basal medium. Change media every 3-4 days for 21 days.
- Qualitative Analysis (Alizarin Red S Staining): a. At day 21, wash scaffolds in PBS and fix in 4% PFA for 30 min. b. Wash with distilled water and incubate with 2% Alizarin Red S solution (pH 4.2) for 20 min. c. Wash extensively with water to remove non-specific stain. Image under brightfield microscope.
- Quantitative Analysis (qPCR for Osteogenic Markers): a. At intermediate (7, 14 days) and endpoint (21 days) time points, homogenize scaffolds in TRIzol to extract total RNA. b. Synthesize cDNA. c. Perform qPCR using primers for early (RUNX2) and late (OSTERIX, OCN) osteogenic markers. Normalize to housekeeping genes (e.g., GAPDH). Analyze via the 2^(-ΔΔCt) method.
Data Presentation
Table 2: Representative Quantitative Data from Scaffold Validation (Hypothetical Data for hMSCs)
| Parameter | Day 1 | Day 7 | Day 14 | Day 21 (Osteogenic) | Notes/Method |
|---|---|---|---|---|---|
| Seeding Efficiency (%) | 85.2 ± 4.1 | - | - | - | DNA Quantification |
| Metabolic Activity (RFU) | 1000 ± 150 | 3200 ± 410 | 6500 ± 720 | 9800 ± 850 | PrestoBlue Assay |
| Total DNA (ng/scaffold) | 105.5 ± 12.3 | 280.7 ± 30.1 | 510.2 ± 45.8 | 750.4 ± 65.5 | PicoGreen Assay |
| ALP Activity (nmol/min/µg DNA) | 5.1 ± 0.8 | 22.5 ± 3.2 | 45.6 ± 5.1 | 18.2 ± 2.5* | Early Osteogenic Marker |
| Calcium Deposition (Absorbance) | 0.05 ± 0.01 | 0.12 ± 0.03 | 0.45 ± 0.08 | 1.85 ± 0.21 | Alizarin Red S Elution |
| RUNX2 Gene Expression (Fold Change) | 1.0 ± 0.2 | 3.5 ± 0.6 | 6.8 ± 1.1 | 15.2 ± 2.3 | qPCR vs. Day 1 Control |
*ALP activity often peaks at intermediate stages of osteogenesis and declines as mineralization intensifies.
Visualizations
Title: Workflow for 3D Scaffold Biological Validation
Title: Key Signaling in Osteogenic Differentiation
Application Notes
Context within 3D Bioprinted Porous Biomimetic Scaffolds Research
The successful translation of 3D-bioprinted scaffolds into clinical applications is critically dependent on their performance following implantation. This "In Vivo Performance" triad—Host Integration, Vascularization, and Functional Outcomes—serves as the ultimate validation metric for scaffold design, biomaterial selection, and biofabrication technique efficacy. Within the thesis on advanced bioprinting, this section provides the essential bridge between in vitro characterization and preclinical utility, evaluating how scaffold architecture (pore size, interconnectivity, mechanical properties) and biochemical functionalization direct biological fate in a living system.
Key Performance Indicators (KPIs) and Quantitative Benchmarks
The following tables summarize critical quantitative metrics for evaluating in vivo performance, derived from recent literature.
Table 1: Metrics for Host Integration & Immunomodulation
| Metric | Assessment Method | Target/Optimal Range (in Early Phase) | Significance |
|---|---|---|---|
| Foreign Body Response (FBR) Capsule Thickness | Histomorphometry (H&E) | < 50-100 µm at 2-4 weeks | Indicates degree of chronic inflammation; thinner capsule suggests better biocompatibility. |
| Macrophage Polarization Ratio (M2:M1) | Immunohistochemistry (CD86/CCR7 vs CD206/ARG1) | M2:M1 > 2.0 at 7-14 days | High M2 phenotype promotes constructive remodeling and integration. |
| Extracellular Matrix (ECM) Infiltration Depth | Masson's Trichrome/ Picrosirius Red | > 75% scaffold depth by 4 weeks | Demonstrates host cell migration and scaffold bioactivity. |
| Degradation Rate Match | μCT/Residual Mass Measurement | Matches tissue ingrowth rate (e.g., 10-30% loss/month) | Prevents mechanical instability or space-filler collapse. |
Table 2: Metrics for Vascularization
| Metric | Assessment Method | Target/Optimal Range | Significance |
|---|---|---|---|
| Perfusion Onset Time | Laser Speckle Contrast Imaging (LSCI), Doppler Ultrasound | Detectable flow within 7-14 days | Indicates rapid anastomosis of new vessels. |
| Vessel Density (within scaffold) | IHC (CD31, α-SMA) | > 200 vessels/mm² at 4 weeks | Critical for nutrient/waste exchange and cell survival in thick scaffolds. |
| Vessel Maturity Index | IHC (α-SMA+ vessels / CD31+ vessels) | > 0.6 at 4-8 weeks | Proportion of stabilized, perfusable vessels vs. total capillaries. |
| Functional Perfusion Volume | Microfil perfusion + μCT | > 40% scaffold volume by 4 weeks | Gold-standard for 3D quantification of connected, blood-carrying network. |
Table 3: Metrics for Functional Outcomes (Bone Tissue Example)
| Metric | Assessment Method | Target/Optimal Range | Significance |
|---|---|---|---|
| New Bone Volume (BV/TV) | μCT Analysis | > 25% at 8-12 weeks | Primary measure of osteogenic functional outcome. |
| Bone-Material Contact (BMC) | Histomorphometry (Toluidine Blue) | > 60% at implant interface | Direct measure of scaffold osteointegration. |
| Compressive Strength Recovery | Mechanical Testing (ex vivo) | > 70% of native tissue strength at 12 weeks | Restoration of biomechanical function. |
| Innervation Density | IHC (PGP9.5, β-III Tubulin) | Presence within new tissue by 8-12 weeks | Indicator of complex, functional tissue regeneration. |
Detailed Experimental Protocols
Protocol: Longitudinal Assessment of Host Integration and Vascularization in a Rodent Subcutaneous Implantation Model
Aim: To quantitatively evaluate the kinetics of immune response, fibrous capsule formation, and de novo vascular network formation in a 3D-bioprinted porous scaffold.
Materials:
- Test Article: Sterilized (gamma-irradiated or EtO) 3D-bioprinted porous scaffold (e.g., 5mm diameter x 2mm thick disc).
- Animal Model: Immunocompetent mouse (e.g., C57BL/6) or rat.
- Anesthesia: Isoflurane/O₂ mixture.
- Analgesia: Buprenorphine SR.
- Surgical Tools: Sterile forceps, scissors, needle holder, 5-0 Vicryl suture, skin stapler.
- In Vivo Imaging System: Laser Speckle Contrast Imager (LSCI) or similar.
- Perfusion & Fixation: Heparinized saline, 4% Paraformaldehyde (PFA), Microfil MV-122 (Flow Tech).
- Ex Vivo Analysis: μCT scanner, histology equipment.
Procedure:
Day 0: Implantation Surgery
- Anesthetize animal. Administer pre-operative analgesic.
- Shave and aseptically prepare the dorsal skin.
- Make a 1cm midline incision. Create two subcutaneous pockets laterally using blunt dissection.
- Implant one scaffold per pocket. For a controlled study, implant a comparator material (e.g., non-porous film) in the contralateral pocket.
- Close the incision with sutures or staples.
- Monitor animal until fully recovered.
Timepoints: 3, 7, 14, 28, 56 days post-implantation (n=5-6/group/timepoint)
- In Vivo Perfusion Imaging (Day 7, 14, 28):
- Anesthetize animal. Depilate the dorsal skin.
- Place animal under LSCI camera. Acquire speckle contrast images over the scaffold region.
- Use software to calculate perfusion units (PU) within a defined ROI over the scaffold versus adjacent native tissue. Calculate relative perfusion.
- Terminal Procedure & Vessel Perfusion Casting (Selected timepoints: 14, 28, 56 days):
- Deeply anesthetize animal.
- Inject Heparin (100 U) via intracardiac route.
- Cannulate the descending aorta. Perfuse with ~50mL heparinized saline at 100 mmHg until effluent is clear.
- Perfuse with 10-15mL of prepared Microfil compound (mixed per manufacturer's instructions).
- Place carcass at 4°C for 1 hour to polymerize the Microfil.
- Explant the scaffold with surrounding tissue and fix in 4% PFA for 48h.
- μCT Analysis of Vascular Cast:
- Scan fixed samples at high resolution (e.g., 10μm isotropic voxels).
- Reconstruct 3D model. Apply global threshold to segment the Microfil signal.
- Calculate: Vessel Volume Fraction (VV/Scaffold Vol), Vessel Thickness Distribution, Connectivity Density.
- Histological Processing & Staining:
- Decalcify if necessary. Dehydrate, paraffin-embed.
- Section at 5μm thickness.
- Stain: H&E (general morphology), Masson's Trichrome (collagen/ECM), Picrosirius Red (collagen birefringence under polarized light), IHC for CD31 (endothelial cells), α-SMA (mature vessels, myofibroblasts), CD68/CD206/CD86 (macrophages).
- Histomorphometry:
- Using image analysis software (e.g., ImageJ, QuPath):
- Measure fibrous capsule thickness at 4-8 equidistant points.
- Count CD31+/α-SMA+ objects to calculate vessel density and maturity index.
- Determine percentage area of stained tissue (e.g., collagen, new bone) within the scaffold pores.
- Using image analysis software (e.g., ImageJ, QuPath):
Protocol: Functional Bone Regeneration in a Critical-Sized Calvarial Defect Model
Aim: To assess the functional outcome of a 3D-bioprinted osteogenic scaffold in restoring bone structure and mechanics.
Materials:
- Test Article: Sterilized, osteoinductive/osteoconductive 3D-bioprinted scaffold, sized to fit defect.
- Animal Model: Rat (e.g., Sprague Dawley) or mouse.
- Defect Creation: Trephine bur (e.g., 5mm diameter for rat), surgical drill.
- Fixation Method: Biocompatible tissue adhesive or very fine sutures.
- In Vivo Imaging: In vivo μCT scanner.
- Ex Vivo Analysis: μCT, mechanical tester, histology.
Procedure:
- Surgery: Anesthetize and prepare the animal. Make a sagittal incision over the cranium. Reflect the periosteum. Create a bilateral or single critical-sized calvarial defect using a trephine bur under constant saline irrigation. Avoid damaging the dura mater.
- Implantation: Implant the test scaffold into the defect. Secure if necessary. Close the wound.
- Longitudinal In Vivo μCT (0, 4, 8, 12 weeks): Anesthetize animal and scan at high resolution. Co-register scans over time to quantify mineralized tissue volume (Bone Volume/Total Volume - BV/TV) within the defect site.
- Termination & Ex Vivo Analysis: At terminal timepoint (e.g., 12 weeks), euthanize and explant the calvarium.
- μCT: Perform high-resolution scan for detailed microarchitectural analysis (Trabecular Number, Thickness, Connectivity Density).
- Biomechanical Testing: Perform a push-out test or compression test using a flat platen on an instron machine to measure failure load and stiffness of the regenerated tissue/scaffold complex.
- Histology: Process for undecalcified (methylmethacrylate embedding) sections if highly mineralized. Perform Goldner's Trichrome or von Kossa/van Gieson staining to distinguish mineralized bone (green/black) from osteoid (red) and quantify Bone-Material Contact.
Visualizations
Diagram Title: In Vivo Performance Pathway Logic
Diagram Title: In Vivo Evaluation Timeline & Assays
The Scientist's Toolkit: Research Reagent Solutions
| Item Name / Category | Supplier Examples | Function & Application |
|---|---|---|
| Microfil MV-122 (Silicon Rubber) | Flow Tech, Inc. | Creates a radio-opaque polymer cast of the entire functional vasculature for high-resolution 3D μCT quantification. |
| Laser Speckle Contrast Imaging (LSCI) Systems | Perimed AB, Moor Instruments | Provides real-time, label-free 2D maps of superficial blood flow for longitudinal perfusion monitoring. |
| CD31/PECAM-1 Antibody | Abcam, Cell Signaling, R&D Systems | Immunohistochemistry marker for pan-endothelial cells, used to quantify total vessel density. |
| α-Smooth Muscle Actin (α-SMA) Antibody | Sigma-Aldrich, Abcam | Marks pericytes and vascular smooth muscle cells; co-staining with CD31 identifies mature, stabilized vessels. |
| Arg1 (Arginase-1) & iNOS Antibodies | Cell Signaling, Santa Cruz | Key markers for M2 (pro-remodeling) and M1 (pro-inflammatory) macrophage polarization, respectively. |
| Osteocalcin (OCN) Antibody | Takara, Abcam | Specific immunohistochemistry marker for mature osteoblasts and newly formed, mineralizing bone. |
| Parafilm-Embedding for Undecalcified Bone | Various | Methylmethacrylate-based embedding preserves mineral content for staining of mineralized tissue in bone regeneration studies. |
| In Vivo μCT Imaging Systems | Bruker, Scanco Medical | Enables longitudinal, non-destructive 3D quantification of scaffold degradation, mineralization (BV/TV), and vascular casts. |
| Picrosirius Red Stain Kit | Sigma-Aldrich, Abcam | Stains collagen fibers; viewed under polarized light, distinguishes mature (thick, red/yellow) from immature (thin, green) collagen. |
This document, framed within a thesis on 3D bioprinting techniques for porous biomimetic scaffolds, provides a comparative analysis of dominant bioprinting modalities. It details application notes, experimental protocols, and research toolkits to guide researchers, scientists, and drug development professionals in selecting and implementing appropriate techniques for specific tissue engineering and regenerative medicine applications.
Bioprinting Modalities: Comparative Analysis
Extrusion-Based Bioprinting
Strengths: High versatility in bioink materials (hydrogels, cell spheroids, thermoplastic polymers); ability to fabricate large, high-density cellular constructs; excellent mechanical integrity of deposited strands; relatively high printing speed and scalability; low cost. Limitations: Limited printing resolution (typically > 100 µm); potential for high shear stress during extrusion, affecting cell viability; nozzle clogging risks; surface roughness of printed filaments.
Inkjet-Based Bioprinting
Strengths: High printing speed and relatively low cost; good resolution (50-300 µm); high cell viability due to low shear stress; capability for precise droplet placement and digital control. Limitations: Limited bioink viscosity range (typically 3.5-12 mPa·s); frequent nozzle clogging; inconsistent droplet generation; difficulty forming mechanically robust structures; limited cell density.
Laser-Assisted Bioprinting (LAB)
Strengths: Nozzle-free, non-contact process eliminating clogging; ultra-high resolution (cell-scale, ~10-50 µm); extremely high cell viability (>95%); ability to print high-viscosity and high-cell-density bioinks. Limitations: Low printing speed and throughput; high cost and system complexity; limited scalability for large constructs; potential for metal nanoparticle contamination from the energy-absorbing layer.
Stereolithography (SLA) / Digital Light Processing (DLP) Bioprinting
Strengths: Excellent resolution (25-100 µm) and surface finish; high architectural accuracy for complex porous geometries; fast printing speed for entire layers simultaneously; good mechanical properties. Limitations: Limited to photo-crosslinkable materials; potential cytotoxicity from photoinitiators and residual monomers; lack of cell-friendly printing environment (often require post-seeding); light scattering can affect depth resolution.
Table 1: Comparative Technical Specifications of Bioprinting Modalities
| Modality | Typical Resolution (µm) | Typical Viscosity Range (mPa·s) | Max Cell Density (cells/mL) | Cell Viability (%) | Relative Speed | Relative Cost |
|---|---|---|---|---|---|---|
| Extrusion | 100 - 500 | 30 - 6x10⁷ | >10⁸ | 40 - 95 | Medium | Low-Medium |
| Inkjet | 50 - 300 | 3.5 - 12 | ~10⁶ | 75 - 90 | High | Low |
| Laser-Assisted | 10 - 50 | 1 - 300 | >10⁸ | 85 - 99 | Low | Very High |
| SLA/DLP | 25 - 100 | Photo-sensitive | Post-seeding | N/A (post-seed) | High | Medium-High |
Table 2: Suitability for Scaffold Properties
| Modality | Pore Size Control | Interconnectivity Fidelity | Mechanical Strength | Biomimetic Complexity | Multi-material Capability |
|---|---|---|---|---|---|
| Extrusion | Good | Good | Excellent | Good | Good (co-axial, multi-head) |
| Inkjet | Fair | Fair | Poor | Fair | Fair (multi-head) |
| Laser-Assisted | Excellent | Excellent | Poor-Fair | Excellent | Fair |
| SLA/DLP | Excellent | Excellent | Good-Excellent | Excellent | Limited (mostly sequential) |
Detailed Experimental Protocols
Protocol 4.1: Extrusion Bioprinting of Alginate-Gelatin Porous Scaffold
Objective: To fabricate a 3D porous, biomimetic scaffold using a thermo-responsive alginate-gelatin composite bioink. Materials: See "Scientist's Toolkit" (Section 6). Procedure:
- Bioink Preparation: Dissolve 4% (w/v) sodium alginate and 8% (w/v) gelatin in DPBS at 37°C under sterile conditions. Filter sterilize (0.22 µm). Mix with human mesenchymal stem cells (hMSCs) at a density of 5x10⁶ cells/mL. Keep at 28°C in printing cartridge to prevent gelation.
- Printer Setup: Load bioink into a temperature-controlled (28°C) syringe. Fit a conical nozzle (22G, 410 µm inner diameter). Set print bed temperature to 15°C.
- Printing Parameters: Set printing pressure to 60-80 kPa, speed to 8 mm/s. Use a 0/90° lay-down pattern with 1.5 mm strand spacing to create a 10-layer scaffold (10x10x3 mm).
- Crosslinking: Post-print, immerse scaffold in 100 mM CaCl₂ solution for 5 mins for ionic crosslinking of alginate.
- Assessment: Assess scaffold morphology via SEM, cell viability via Live/Dead assay at 1 and 7 days.
Protocol 4.2: DLP Bioprinting of Methacrylated Gelatin (GelMA) Lattice Scaffold
Objective: To create a high-resolution, photocrosslinked porous lattice scaffold for cell seeding studies. Materials: See "Scientist's Toolkit" (Section 6). Procedure:
- Bio-resin Preparation: Synthesize GelMA (10% w/v) and dissolve in DPBS containing 0.5% (w/v) lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) photoinitiator. Sterilize via filtration.
- Digital Design: Create a 3D model of a gyroid lattice (pore size 300 µm, porosity 70%) using CAD software. Slice into 50 µm layers.
- Printing: Pour bio-resin into the DLP printer vat. Set exposure time per layer to 8 seconds (405 nm light, 20 mW/cm²). Print the scaffold layer-by-layer.
- Post-processing: Rinse printed scaffold in sterile PBS to remove uncured resin. Perform secondary crosslinking under UV light (365 nm, 5 mins) for enhanced mechanical properties.
- Cell Seeding: Seed the sterilized scaffold with NIH/3T3 fibroblasts at 10⁵ cells/scaffold using a centrifugal seeding method (1000 rpm for 5 mins).
- Culture & Analysis: Culture in DMEM for up to 14 days. Analyze cell proliferation (Alamar Blue) and infiltration (histology) at days 1, 7, and 14.
Visualization Diagrams
Diagram Title: Extrusion Bioprinting Workflow
Diagram Title: DLP/SLA Photopolymerization Pathway
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Featured Protocols
| Material/Reagent | Function & Rationale | Example Vendor/Product |
|---|---|---|
| Sodium Alginate (High G-Content) | Provides biocompatibility and rapid ionic crosslinking capability for shape fidelity. | Sigma-Aldrich, 71238 |
| Gelatin (Type A, from porcine skin) | Provides cell-adhesive RGD motifs and thermo-responsive behavior for printability. | Sigma-Aldrich, G2500 |
| GelMA (Methacrylated Gelatin) | Photocrosslinkable hydrogel backbone combining bioactivity with tunable mechanics. | Advanced BioMatrix, Gelin-S-MA |
| Lithium Phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Cytocompatible photoinitiator with high absorption at 405 nm for rapid crosslinking. | Sigma-Aldrich, 900889 |
| Calcium Chloride (CaCl₂) Solution | Ionic crosslinker for alginate, forming stable "egg-box" structures. | Thermo Fisher Scientific |
| hMSCs (Human Mesenchymal Stem Cells) | Model multipotent cell type for osteogenic/chondrogenic differentiation studies in scaffolds. | Lonza, PT-2501 |
| Fluorescent Live/Dead Viability Kit (Calcein AM/EthD-1) | Two-color fluorescence assay to quantify live (green) and dead (red) cells in 3D constructs. | Thermo Fisher, L3224 |
| Cell Culture-Tested Dimethyl Sulfoxide (DMSO) | Cryoprotectant for cell storage and solvent for certain biocompatible dyes. | Sigma-Aldrich, D2650 |
Within the broader thesis on 3D bioprinting techniques for porous biomimetic scaffolds, this application note presents a comparative case study. It evaluates two dominant fabrication paradigms—extrusion-based bioprinting and light-based (vat polymerization) bioprinting—for generating scaffolds intended to promote osteogenesis (bone formation). The comparison focuses on architectural, mechanical, and biological outcome parameters critical for bone tissue engineering.
Table 1: Architectural & Mechanical Properties Comparison
| Parameter | Extruded Scaffolds | Light-Printed Scaffolds | Measurement Method |
|---|---|---|---|
| Feature Resolution | 100 - 500 µm | 10 - 200 µm | Microscopy (SEM) |
| Average Pore Size | 200 - 800 µm | 100 - 500 µm | Micro-CT analysis |
| Porosity | 50 - 70% | 60 - 85% | Micro-CT / Gravimetry |
| Compressive Modulus | 1 - 50 MPa | 0.5 - 10 MPa | Mechanical testing |
| Print Speed | 1 - 10 mm³/s | 5 - 100 mm³/s | Printer settings |
| Common Materials | Alginate, GelMA, PLA, PCL, Bio-inks with ceramics | PEGDA, GelMA, HA-nanocomposite resins | Material formulation |
Table 2: In Vitro Osteogenic Performance Summary
| Outcome Metric | Extruded Scaffolds | Light-Printed Scaffolds | Assay & Timepoint |
|---|---|---|---|
| Cell Viability (Day 1) | 75-90% | 85-98% | Live/Dead staining |
| Cell Proliferation (Day 7) | Moderate (1.5-2x increase) | High (2-3x increase) | AlamarBlue/CCK-8 |
| ALP Activity (Day 14) | Moderate | High | ALP enzymatic assay |
| Calcium Deposition (Day 21) | Significant | Highly Significant | Alizarin Red S staining |
| Osteogenic Gene (Runx2) Upregulation | 3-5 fold | 5-10 fold | qPCR (Day 14) |
Experimental Protocols
Protocol 1: Fabrication of Extruded Scaffolds (GelMA/Hydroxyapatite Composite)
- Objective: To create porous, osteoconductive scaffolds via pneumatic extrusion.
- Materials: Methacrylated gelatin (GelMA, 10% w/v), nano-hydroxyapatite (nHA, 5% w/v), LAP photoinitiator (0.25% w/v), sterile PBS.
- Procedure:
- Bioink Preparation: Dissolve GelMA in PBS at 40°C. Mix in nHA powder uniformly using a dual-syringe mixer. Add LAP and protect from light.
- Printing: Load bioink into a sterile syringe. Use a 22G conical nozzle. Set pneumatic pressure (20-35 kPa) and print speed (8 mm/s) for consistent fiber deposition.
- Crosslinking: Print in a 0-4°C cold stage to maintain viscosity. Immediately after printing, expose the scaffold to 405 nm UV light (5 mW/cm²) for 60 seconds per layer.
- Post-processing: Wash scaffolds 3x in PBS to remove uncrosslinked material. Sterilize under UV light for 30 minutes.
Protocol 2: Fabrication of Light-Printed Scaffolds (Digital Light Processing - DLP)
- Objective: To fabricate high-resolution scaffolds using a digital micromirror device.
- Materials: PEGDA (MW 700 Da, 30% v/v), GelMA (5% w/v), nHA (2% w/v), LAP (0.3% w/v).
- Procedure:
- Resin Preparation: Combine PEGDA, GelMA, and nHA in PBS. Sonicate for 30 min to disperse nHA. Add LAP and mix in the dark until fully dissolved. Filter sterilize (0.22 µm).
- Printing: Load resin into the DLP printer vat. Use a sliced 3D model (e.g., gyroid lattice, 300 µm pore size). Set layer thickness to 50 µm and exposure time to 3 seconds per layer (405 nm light, 10 mW/cm²).
- Post-processing: Carefully remove printed scaffold. Rinse in 70% ethanol for 2 min to remove uncured resin, then wash 3x in PBS. Perform a secondary UV crosslink (365 nm, 5 min) for enhanced stability.
Protocol 3: Standardized In Vitro Osteogenesis Assay
- Objective: To evaluate and compare scaffold osteoinductivity.
- Cell Culture: Seed human mesenchymal stem cells (hMSCs) on sterilized scaffolds at 50,000 cells/scaffold. Maintain in growth medium (GM) for 24h, then switch half to osteogenic medium (OM: GM + 10 mM β-glycerophosphate, 50 µg/mL ascorbic acid, 100 nM dexamethasone).
- Assessment Timepoints:
- Day 1/3/7: Cell viability (Live/Dead) and proliferation (AlamarBlue).
- Day 7/14: Alkaline Phosphatase (ALP) activity (pNPP assay) and early osteogenic gene expression (Runx2, ALPL via qPCR).
- Day 21/28: Calcium mineralization (Alizarin Red S quantification at 562 nm).
Signaling Pathways in Scaffold-Mediated Osteogenesis
Diagram Title: Scaffold Properties Activate Osteogenic Signaling Pathways
Experimental Workflow for Comparative Study
Diagram Title: Comparative Scaffold Study Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
| Item Name | Function in Experiment | Example/Catalog Consideration |
|---|---|---|
| Methacrylated Gelatin (GelMA) | Photocrosslinkable bioink base providing cell-adhesive RGD motifs. | Sigma-Aldrich 900637, Advanced BioMatrix GME-30-RGD |
| Poly(ethylene glycol) diacrylate (PEGDA) | Biocompatible, tunable resin for high-resolution light printing. | Sigma-Aldrich 701963 |
| Nano-Hydroxyapatite (nHA) | Osteoconductive ceramic mimicking bone mineral content. | Berkeley Advanced Biomat. Inc., Sigma 677418 |
| Lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) | Efficient, cytocompatible photoinitiator for UV/blue light crosslinking. | Tokyo Chemical Industry L0231 |
| Human Mesenchymal Stem Cells (hMSCs) | Primary cell model for studying osteogenic differentiation. | Lonza PT-2501, ATCC PCS-500-012 |
| Osteogenic Supplement Kit | Provides dexamethasone, ascorbate, and β-glycerophosphate for differentiation. | Gibco A1007201 |
| AlamarBlue Cell Viability Reagent | Fluorescent resazurin-based assay for quantifying proliferation. | Invitrogen DAL1100 |
| Alizarin Red S Solution | Dye that binds calcium deposits for quantifying mineralization. | ScienCell ACS-301 |
| qPCR Probes for Runx2 & ALPL | Primers/probes for quantifying osteogenic gene expression. | TaqMan Assays (Thermo Fisher) |
Conclusion
The convergence of advanced 3D bioprinting techniques with sophisticated biomaterial science is revolutionizing the fabrication of porous, biomimetic scaffolds. From foundational design principles to validated clinical potential, this field requires a balanced approach that prioritizes both structural fidelity and biological functionality. The future lies in developing intelligent, multi-material bioinks, integrating real-time monitoring sensors, and creating patient-specific constructs via clinical imaging data. For researchers and drug developers, mastering these techniques is pivotal for advancing regenerative therapies and developing highly predictive human-relevant models, ultimately accelerating the translation of lab-based innovations into tangible clinical solutions.