Nanoparticle-Mediated CRISPR-Cas9 RNP Delivery: A Comprehensive Guide for Therapeutic Development
This article provides a detailed exploration of nanoparticle (NP)-based delivery systems for CRISPR-Cas9 ribonucleoprotein (RNP) complexes.
Nanoparticle-Mediated CRISPR-Cas9 RNP Delivery: A Comprehensive Guide for Therapeutic Development
Abstract
This article provides a detailed exploration of nanoparticle (NP)-based delivery systems for CRISPR-Cas9 ribonucleoprotein (RNP) complexes. Targeted at researchers and drug development professionals, it covers the foundational rationale for RNP delivery over DNA-based methods, surveys current nanoparticle platforms (lipidic, polymeric, inorganic), and details key formulation and characterization methodologies. It addresses common challenges in stability, cellular uptake, endosomal escape, and cargo release, offering optimization strategies. The content further validates approaches through comparative analysis of delivery efficiency, specificity, and safety profiles across NP classes, concluding with an outlook on clinical translation and next-generation nanocarriers for genome editing therapeutics.
Why RNP Delivery? Understanding the Core Advantages and Nanoparticle Imperative
This Application Note delineates the advantages and methodologies for employing the CRISPR-Cas9 Ribonucleoprotein (RNP) complex as a transient, DNA-free genome editing tool. Positioned within a broader thesis on nanoparticle-mediated RNP delivery, this document underscores the critical importance of the RNP format for enhancing safety profiles and reducing off-target effects in therapeutic development. The defined, transient nature of the RNP complex circumvents risks associated with prolonged nuclease expression from DNA templates, making it the optimal payload for advanced nanoparticle delivery systems.
Comparative Analysis of CRISPR Delivery Formats
The following table summarizes the core quantitative attributes distinguishing RNP delivery from plasmid DNA (pDNA) and messenger RNA (mRNA) based methods.
Table 1: Quantitative Comparison of CRISPR-Cas9 Delivery Modalities
| Feature | Plasmid DNA (pDNA) | mRNA + sgRNA | RNP Complex |
|---|---|---|---|
| Time to Active Nuclease (hr) | 24 - 72 (requires transcription/translation) | 2 - 12 (requires translation) | < 1 (immediately active) |
| Duration of Nuclease Activity | Prolonged (days) | Moderate (hours to days) | Short (hours) |
| Off-Target Effect Incidence | High | Moderate | Low |
| Risk of Genomic Integration | Yes (random integration) | No | No |
| Immunogenicity Risk | High (bacterial sequences, TLR9) | High (modified nucleotides reduce risk) | Low (protein/sRNA) |
| Primary Editing Outcome | NHEJ/HDR | NHEJ/HDR | Predominantly NHEJ |
| Optimal for Nanoparticle Delivery | Poor (large size, complex unpacking) | Good (moderate size) | Excellent (defined complex size) |
Protocol 1: In Vitro Assembly and Validation of Cas9-sgRNA RNP
Objective: To assemble, purify, and validate functional Cas9 RNP complexes for downstream delivery experiments.
Materials (Research Reagent Solutions):
- Purified Cas9 Nuclease: Recombinant S. pyogenes Cas9 protein, endotoxin-free. Function: DNA cleavage effector.
- Synthetic sgRNA: Chemically modified, HPLC-purified single-guide RNA. Function: Targets Cas9 to specific genomic locus.
- Nuclease-Free Duplex Buffer (IDT): 30 mM HEPES, 100 mM potassium acetate. Function: Optimal ionic conditions for RNP assembly.
- Electrophoretic Mobility Shift Assay (EMSA) Gel: 6% native polyacrylamide gel. Function: Visualizes RNP complex formation.
- In Vitro Cleavage Assay Substrate: PCR-amplified DNA target (~500 bp). Function: Validates RNP enzymatic activity.
Methodology:
- RNP Assembly: Combine 10 pmol of purified Cas9 protein with 12 pmol of sgRNA (1:1.2 molar ratio) in duplex buffer. Final volume: 20 µL.
- Incubation: Incubate mixture at 25°C for 10 minutes to allow complex formation.
- EMSA Validation: Load 5 µL of the assembly reaction on a pre-chilled 6% native PAGE gel. Run at 100V for 60 min in 0.5x TBE buffer at 4°C. Stain with SYBR Gold. A successful shift (Cas9+sgRNA vs. Cas9 alone) confirms complex formation.
- In Vitro Cleavage Assay: Add 100 ng of target DNA substrate to 5 µL of the assembled RNP. Incubate at 37°C for 60 min in 1x Cas9 reaction buffer. Heat-inactivate at 70°C for 10 min. Analyze products on a 2% agarose gel. Cleavage is indicated by the appearance of two lower molecular weight bands.
Title: RNP Assembly and Validation Workflow
Protocol 2: Assessing RNP Delivery & Editing Efficiency via Nanoparticle Formulation
Objective: To formulate lipid nanoparticles (LNPs) encapsulating pre-assembled RNP and quantify cellular editing efficiency and kinetics.
Materials (Research Reagent Solutions):
- Ionizable Cationic Lipid (e.g., DLin-MC3-DMA): Function: LNP core component, enables endosomal escape.
- PEGylated Lipid: Function: Stabilizes LNP surface and modulates pharmacokinetics.
- Microfluidic Mixer (e.g., NanoAssemblr): Function: Enables reproducible, rapid LNP formulation.
- HEK293T Cells with GFP Reporter: Stably expressing GFP with an in-frame stop codon targeted by sgRNA. Function: Quantitative editing readout via flow cytometry (GFP recovery).
- T7 Endonuclease I (T7E1) or ICE Analysis Software: Function: Detects indels at endogenous genomic loci.
Methodology:
- LNP Formulation: Prepare an ethanol phase containing ionizable lipid, phospholipid, cholesterol, and PEG-lipid. Prepare an aqueous phase containing assembled RNP in citrate buffer (pH 4.0). Use a microfluidic mixer to combine phases at a 3:1 ratio (aqueous:ethanol). Dialyze against PBS to remove ethanol and raise pH.
- LNP Characterization: Use dynamic light scattering (DLS) to measure particle size and PDI. Use RiboGreen assay to measure encapsulation efficiency (% of RNP protected from RNase).
- Cell Transfection: Seed HEK293T GFP-reporter cells in a 24-well plate. At 80% confluency, treat cells with RNP-LNPs (e.g., 100 nM RNP final concentration). Include controls: naked RNP and untreated cells.
- Efficiency & Kinetics Analysis:
- Flow Cytometry: At 24, 48, 72, and 96 hours post-transfection, harvest cells and analyze GFP-positive population (%) via flow cytometry.
- Genomic Analysis: At 72 hours, extract genomic DNA from treated (non-reporter) cells. PCR-amplify the on-target region. Digest PCR products with T7E1 enzyme and analyze on agarose gel to calculate indel frequency. Use Sanger sequencing and ICE analysis for precise quantification.
Title: RNP-LNP Delivery and Analysis Pathway
The Scientist's Toolkit: Essential Reagents for RNP-Based Editing
Table 2: Key Research Reagent Solutions for RNP Experiments
| Item | Function & Rationale |
|---|---|
| Recombinant Cas9 Protein (WT or HiFi) | The core nuclease. HiFi variants reduce off-target activity, crucial for therapeutic applications. |
| Chemically Modified sgRNA (2'-O-Methyl, Phosphorothioate) | Enhances stability against cellular nucleases, increasing RNP half-life and efficacy. |
| Ionizable Lipid Nanoparticles (LNPs) | The leading delivery vector. Protects RNP, facilitates cellular uptake and endosomal escape. |
| Cell-Penetrating Peptides (CPPs) | Alternative delivery method; can conjugate directly to RNP for simplified formulation. |
| T7 Endonuclease I / Surveyor Nuclease | Accessible tools for initial detection of nuclease-induced indels at target sites. |
| Next-Generation Sequencing (NGS) Library Prep Kits | Gold-standard for unbiased, quantitative assessment of on-target editing and genome-wide off-target screening. |
| RiboGreen/Quant-iT Assay | Fluorometric quantification of nucleic acid encapsulation efficiency within nanoparticles. |
| Microfluidic Mixing Platforms | Enables scalable, reproducible production of monodisperse nanoparticle formulations. |
This application note, framed within ongoing CRISPR-Cas9 ribonucleoprotein (RNP) delivery research, details the key constraints of traditional plasmid and viral vector systems for gene editing. While plasmid and viral methods have been foundational, their immunogenicity, off-target editing profiles, and uncontrolled persistent expression present significant hurdles for therapeutic translation. This document provides quantitative comparisons and protocols for assessing these limitations, with a focus on informing the rationale for nanoparticle-mediated RNP delivery.
Comparative Analysis of Delivery Limitations
Table 1: Quantitative Comparison of Key Delivery Vector Limitations
| Limitation Parameter | Plasmid DNA (Non-Viral) | Adenoviral (AdV) Vector | Adeno-Associated Viral (AAV) Vector |
|---|---|---|---|
| Immunogenicity Trigger | CpG motif-mediated TLR9 signaling; potential anti-dsDNA antibodies. | Strong innate & adaptive immune response to viral capsid proteins; pre-existing immunity in >90% of adults. | Capsid-specific T-cell response; neutralizing antibodies (NAb) in 30-70% of population. |
| Off-Target Effect Rate | Higher potential due to prolonged Cas9 expression. Studies show up to 10-fold increase vs. RNP in some cell lines. | Variable; sustained expression can increase risk. | High concern; persistent expression can lead to chronic genotoxicity. Baseline rates vary by serotype & target. |
| Expression Kinetics | Onset: 6-24h; Duration: Days to weeks (transient) but can integrate randomly. | Onset: Rapid (24-48h); Duration: Weeks (episomal, non-integrating). | Onset: Slow (days); Duration: Persistent (months to years, episomal). |
| Typical Payload | DNA encoding Cas9 & gRNA. | DNA encoding Cas9 & gRNA (~8 kb capacity). | DNA encoding Cas9 & gRNA (~4.7 kb capacity, limiting full SpCas9 delivery). |
| Clinical Challenge Example | Inflammatory responses in in vivo delivery; low efficiency. | Severe inflammatory cytokine storms in early trials (e.g., Jesse Gelsinger case). | Fatal SAE in X-linked myotubular myopathy trial (AT132) linked to high-dose AAV and hepatotoxicity. |
Detailed Protocols for Assessing Limitations
Protocol 1: Assessing Immunogenicity of Viral Vectors viaIn VitroImmune Cell Activation Assay
Objective: To quantify innate immune activation (e.g., cytokine release) by viral vectors in human peripheral blood mononuclear cells (PBMCs).
Research Reagent Solutions:
- Fresh or Cryopreserved Human PBMCs: Source of primary immune cells (dendritic cells, monocytes).
- Viral Vectors (AAV, AdV) & Control Plasmid: Test articles at clinical-grade purity.
- LPS (Lipopolysaccharide) & Poly(I:C): Positive controls for TLR4 and TLR3 signaling, respectively.
- Cell Culture Media (RPMI-1640 + 10% FBS): For maintaining PBMC viability.
- Human Cytokine Multiplex Assay Kit (e.g., Luminex): For quantifying IL-6, TNF-α, IFN-α, IFN-γ.
Methodology:
- Isolate PBMCs from healthy donor blood using density gradient centrifugation (Ficoll-Paque).
- Seed 2 x 10^5 PBMCs per well in a 96-well U-bottom plate in 200 µL complete media.
- Treat cells with:
- Test Vectors: AAV (1e10 vg/mL), AdV (1e9 vp/mL), plasmid (1 µg/mL).
- Positive Controls: LPS (100 ng/mL), Poly(I:C) (1 µg/mL).
- Negative Control: Media only.
- Incubate at 37°C, 5% CO2 for 24 hours.
- Centrifuge plate at 300 x g for 5 min. Collect 150 µL of supernatant per well.
- Analyze supernatant using the multiplex cytokine array per manufacturer's protocol.
- Data Analysis: Normalize cytokine levels to media control. A ≥2-fold increase (p<0.05) in pro-inflammatory cytokines (IL-6, TNF-α) indicates significant immunogenicity.
Protocol 2: Quantifying Off-Target Effects via GUIDE-seq for Plasmid vs. RNP Delivery
Objective: To compare genome-wide off-target sites of CRISPR-Cas9 delivered via plasmid vs. RNP format in HEK293T cells.
Research Reagent Solutions:
- SpCas9 Protein: Purified, recombinant.
- Chemically Modified sgRNA or crRNA:tracrRNA duplex: For RNP formation.
- Plasmid Encoding SpCas9 and sgRNA Expression Cassette: (e.g., pX459).
- GUIDE-seq Oligonucleotide (dsODN): Tag for marking double-strand breaks.
- Next-Generation Sequencing (NGS) Library Prep Kit: For GUIDE-seq library construction.
- Transfection Reagent (Lipofectamine): For plasmid delivery. Electroporation device for RNP delivery.
Methodology:
- Cell Preparation: Culture HEK293T cells to 80% confluency in a 6-well plate.
- Delivery:
- RNP Condition: Complex 5 µg Cas9 protein with 200 pmol sgRNA. Add 100 pmol GUIDE-seq dsODN. Deliver via nucleofection.
- Plasmid Condition: Transfect 2 µg pX459 plasmid + 100 pmol GUIDE-seq dsODN using lipofection reagent.
- Harvest genomic DNA 72 hours post-delivery using a silica-column kit.
- Perform GUIDE-seq library preparation as originally described (Tsai et al., Nat Biotechnol, 2015). Briefly: shear DNA, end-repair, A-tail, ligate adaptors, amplify with GUIDE-seq-specific primers.
- Sequence libraries on an Illumina MiSeq (2x150 bp).
- Data Analysis: Use the GUIDE-seq software pipeline to align reads, detect dsODN integration sites, and call off-target loci. Compare the number, location, and frequency of off-target sites between RNP and plasmid conditions.
Protocol 3: Measuring Persistent Expression from AAV Vectors via Luciferase Bioluminescence Imaging
Objective: To monitor the long-term, uncontrolled expression profile of an AAV-delivered transgene in vivo.
Research Reagent Solutions:
- AAV vector encoding Firefly Luciferase (AAV-CB-Luc): Serotype 9 for broad tropism.
- IVIS Spectrum In Vivo Imaging System: For bioluminescent detection.
- D-Luciferin, Potassium Salt: Substrate for firefly luciferase.
- Animal Model (e.g., C57BL/6 mice): For in vivo study.
- Isoflurane Anesthesia System: For animal immobilization during imaging.
Methodology:
- Inject mice intravenously with 1e11 vg of AAV-CB-Luc (n=5) or PBS (n=3 control).
- At scheduled time points (Day 3, 7, 14, 30, 60, 90), inject mice intraperitoneally with 150 mg/kg D-luciferin.
- Anesthetize mice with isoflurane and place in the IVIS imaging chamber 10 minutes post-luciferin injection.
- Acquire bioluminescence images using a standardized exposure time (e.g., 60 seconds).
- Quantify total flux (photons/second) within a fixed region of interest (ROI) using Living Image software.
- Data Analysis: Plot bioluminescence signal over time. Persistent, non-declining signal beyond 60 days indicates stable, long-term expression characteristic of AAV, highlighting the risk of sustained Cas9 activity.
Visualizing Immune Activation Pathways and Experimental Workflow
Diagram Title: Viral Immune Activation Pathway
Diagram Title: GUIDE-seq Workflow for Off-Target Analysis
The application of CRISPR-Cas9 as a pre-assembled ribonucleoprotein (RNP) complex, delivered via engineered nanoparticles, represents a paradigm shift in precision genome editing. Within the broader thesis of nanoparticle-mediated RNP delivery research, this approach circumforms key limitations of DNA-based delivery (e.g., prolonged Cas9 expression, immunogenicity, insertional mutagenesis). The transient nature of RNP activity directly confers the titular advantages: enhanced specificity through reduced exposure time, minimized off-target edits, and rapid cellular turnover that limits unintended genomic alterations.
Table 1: Comparative Performance Metrics of CRISPR-Cas9 Delivery Modalities
| Metric | RNP (Nanoparticle-Delivered) | Plasmid DNA (Transfection) | Source/Reference |
|---|---|---|---|
| On-Target Editing Efficiency (%) | 60-85% (varies by cell type & target) | 40-70% | 1, 2 |
| Off-Target Mutation Frequency (relative) | 0.1 - 0.5x | 1.0x (baseline) | 3, 4 |
| Cas9 Intracellular Persistence | < 24-48 hours | Days to weeks | 5, 6 |
| Indel Pattern Consistency | High | Moderate to Low | 7 |
| Cellular Toxicity (relative) | Low | Moderate to High | 8 |
| Immune Response Activation | Minimal | Significant (anti-Cas9 antibodies) | 9 |
Sources synthesized from recent literature (2022-2024):
- Nature Nanotechnology, 2023, Lipid nanoparticle RNP delivery in vivo.
- Nucleic Acids Research, 2023, Primary T-cell editing.
- Nature Methods, 2022, GUIDE-seq analysis of RNP specificity.
- Cell Reports, 2024, Comparative off-target profiling.
- PNAS, 2023, RNP kinetics study.
- Gene Therapy, 2022, Persistence assay.
- CRISPR Journal, 2023, Sequencing analysis.
- Biomaterials, 2024, Cytotoxicity screening.
- Science Advances, 2023, Immunogenicity profiling.
Key Signaling Pathways & Cellular Processing
Diagram 1: RNP Uptake, Processing, and Turnover Pathway
Detailed Experimental Protocols
Protocol 4.1: Formulation of Cationic Lipid Nanoparticles (LNPs) for RNP Encapsulation
Objective: Prepare stable, serum-resistant LNPs encapsulating CRISPR-Cas9 RNP. Materials: See "Scientist's Toolkit" below. Procedure:
- RNP Complex Formation: Dilute purified recombinant Cas9 protein and synthetic sgRNA in nuclease-free duplex buffer (30 mM HEPES, 100 mM KCl, pH 7.5). Mix at a 1:1.2 molar ratio (Cas9:sgRNA). Incubate at 25°C for 10 min.
- Lipid Mixture Preparation: In ethanol, combine ionizable cationic lipid (e.g., DLin-MC3-DMA, 50 mol%), phospholipid (DSPC, 10 mol%), cholesterol (38.5 mol%), and PEG-lipid (1.5 mol%). Warm to 60°C.
- Aqueous Phase Preparation: Dilute the formed RNP complex in citrate buffer (50 mM, pH 4.0).
- Microfluidic Mixing: Using a microfluidic device (e.g., NanoAssemblr), mix the ethanolic lipid stream with the aqueous RNP stream at a 1:3 volumetric flow rate ratio (total flow rate 12 mL/min). Collect effluent in PBS.
- Buffer Exchange & Purification: Dialyze the collected LNP suspension against 1x PBS (pH 7.4) for 4 hours at 4°C using a 100 kDa MWCO membrane. Concentrate using centrifugal filters (100 kDa MWCO).
- Characterization: Measure particle size and PDI via dynamic light scattering (DLS). Determine encapsulation efficiency using a Quant-iT RiboGreen assay for unencapsulated sgRNA.
Protocol 4.2: Off-Target Assessment via GUIDE-seq
Objective: Quantify genome-wide off-target effects of nanoparticle-delivered RNP vs. plasmid transfection. Materials: GUIDE-seq oligonucleotide duplex, PCR reagents, next-generation sequencing (NGS) platform, TAGMENT enzyme. Procedure:
- Cell Treatment & Transfection: Seed HEK293T cells (or target cell line) in 6-well plates. For test group, treat with RNP-loaded LNPs (Protocol 4.1). For control, transfect with a Cas9/sgRNA expression plasmid. Co-deliver 100 pmol of GUIDE-seq oligonucleotide duplex with each modality.
- Genomic DNA Extraction: Harvest cells 72 hours post-treatment. Extract high-molecular-weight gDNA using a silica-column kit.
- Library Preparation:
- Shear 1.5 µg gDNA to ~500 bp via sonication.
- End-repair, A-tail, and ligate Illumina sequencing adapters.
- Perform GUIDE-seq tag-specific PCR enrichment (15 cycles) using primers containing partial Illumina sequences.
- Follow with a second PCR (12 cycles) to add full indices and sequencing handles.
- Sequencing & Analysis: Pool libraries and sequence on an Illumina MiSeq (2x150 bp). Align reads to the reference genome (hg38). Identify double-strand break sites via detection of integrated GUIDE-seq tag using the published GUIDE-seq computational pipeline. Compare off-target site lists and read counts between RNP and plasmid groups.
Experimental Workflow for RNP-NP Research
Diagram 2: Integrated RNP-Nanoparticle Experiment Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Nanoparticle-Mediated RNP Delivery Research
| Reagent/Material | Supplier Examples | Function in Protocol |
|---|---|---|
| Recombinant S. pyogenes Cas9 Nuclease | Thermo Fisher (TrueCut), IDT, Aldevron | The editing enzyme; high-purity, endotoxin-free protein is critical for RNP assembly and low toxicity. |
| Chemically Modified sgRNA (2'-O-methyl, Phosphorothioate) | Synthego, IDT, Trilink | Enhances nuclease resistance and RNP stability; reduces immune activation. |
| Ionizable Cationic Lipid (e.g., DLin-MC3-DMA) | Avanti Polar Lipids, BroadPharm | Key LNP component for efficient encapsulation and endosomal escape of RNP. |
| PEG-lipid (e.g., DMG-PEG2000) | Avanti Polar Lipids, NOF America | Provides nanoparticle stability, reduces aggregation, and modulates pharmacokinetics. |
| Microfluidic Mixer (NanoAssemblr) | Precision NanoSystems | Enables reproducible, scalable formation of monodisperse RNP-loaded LNPs. |
| RiboGreen Assay Kit | Thermo Fisher | Quantifies sgRNA encapsulation efficiency within nanoparticles. |
| GUIDE-seq Kit | Integrated DNA Technologies (IDT) | All-in-one reagents for genome-wide off-target cleavage profiling. |
| T7 Endonuclease I / ICE Assay | NEB, Synthego | Rapid, initial assessment of on-target editing efficiency. |
| Cell-Penetrating Peptide (CPP) Conjugates | PeptideSpec, CPC Scientific | Alternative delivery strategy for RNP; used for comparative studies. |
| Next-Generation Sequencing Kit (Illumina) | Illumina | Essential for deep sequencing of target loci and off-target sites. |
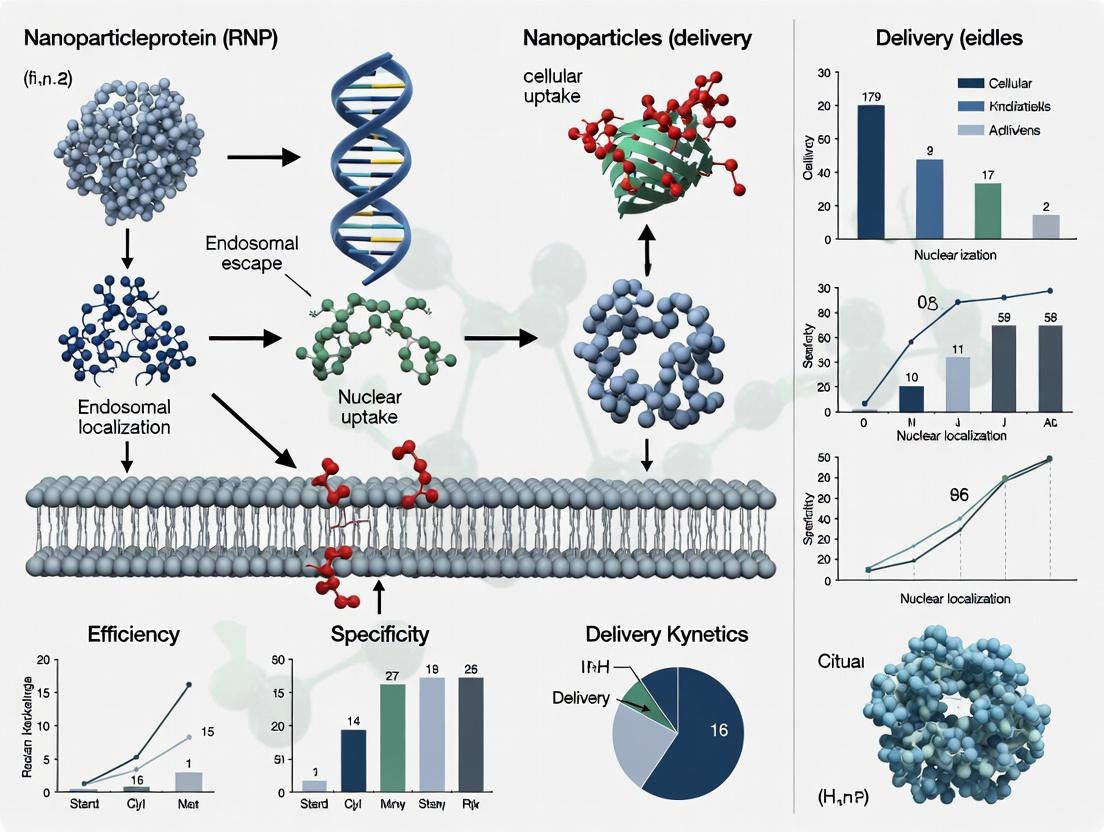
Within the broader context of CRISPR-Cas9 ribonucleoprotein (RNP) delivery via nanoparticles, the final intracellular journey of the RNP complex presents a critical bottleneck. Successful genome editing requires not just cellular uptake, but also endosomal escape, cytosolic stability, and nuclear import of the functional RNP. This Application Note details the primary barriers to intracellular RNP transport and provides protocols for quantitatively assessing these hurdles, enabling the rational design of more effective nanoparticle delivery systems.
Quantitative Analysis of Intracellular Delivery Barriers
The efficiency of each step in the intracellular transport pathway is typically low. The following table summarizes key quantitative benchmarks from recent literature.
Table 1: Typical Efficiency Metrics for Intracellular RNP Delivery Steps
| Delivery Stage | Typical Efficiency Range | Key Measurement Method |
|---|---|---|
| Cellular Uptake | 70-95% of cells show association | Flow cytometry (Cy5-labeled RNP) |
| Endosomal Escape | 1-20% of internalized RNPs | Galectin-8/-9 recruitment assays, split-GFP reporters |
| Cytosolic Availability | <10% of escaped RNPs remain functional | Fluorescence correlation spectroscopy (FCS) |
| Nuclear Import | 1-5% of cytosolic RNPs (in dividing cells) | Nuclear fractionation & immunoblot, live-cell imaging |
| Final Editing Efficiency | 0.1-60% (highly variable by cell type) | NGS, T7E1 assay, flow cytometry for reporter cells |
Table 2: Factors Influencing Cytosolic & Nuclear RNP Transport
| Factor | Impact on RNP Transport | Experimental Modulator |
|---|---|---|
| RNP Size | >40 nm diameter impedes passive nuclear entry | Ultrafiltration, analytical centrifugation |
| NLS Presence/Type | Classical NLS (cNLS) vs. non-classical; can increase nuclear import 5-10 fold | Genetic fusion (e.g., SV40 NLS), chemical conjugation |
| Cell Cycle Phase | Import efficiency increases 3-5x during mitosis/ interphase | Cell cycle synchronization (e.g., thymidine, nocodazole) |
| Cytosolic Nucleases | RNP half-life can be <15 minutes | Co-delivery of nuclease inhibitors (e.g., Aurintricarboxylic acid) |
| Cytosolic Viscosity & Crowding | Diffusion coefficient reduced 2-4x vs. buffer | Microrheology using tracer particles |
Detailed Experimental Protocols
Protocol 1: Quantifying Endosomal Escape Using a Galectin-9-mCherry Reporter Assay
Principle: Cytosolic exposure of glycans (e.g., from ruptured endosomes) recruits the protein Galectin-9. Colocalization of Galectin-9-mCherry puncta with fluorescently labeled RNP indicates failed escape.
Materials:
- HeLa or U2OS cells stably expressing Galectin-9-mCherry.
- Cy5-labeled Cas9 RNP (pre-assembled).
- Nanoparticle delivery formulation.
- Live-cell imaging medium.
- Confocal microscope with environmental chamber.
Procedure:
- Seed Galectin-9-mCherry reporter cells in an 8-chamber glass-bottom dish 24h prior.
- Treat cells with nanoparticle-formulated Cy5-RNP (e.g., 200 nM RNP equivalence).
- At 2, 4, 6, and 8 hours post-treatment, replace medium with live-cell imaging medium.
- Acquire z-stack images (e.g., 3 slices, 0.5 μm interval) for mCherry (ex 587/em 610) and Cy5 (ex 650/em 670) channels.
- Analysis: Use ImageJ/Fiji with coloc2 plugin. Threshold images. Calculate the percentage of Cy5-positive vesicles that colocalize with Galectin-9-mCherry puncta. A decrease in colocalization over time suggests successful escape.
Protocol 2: Assessing Nuclear Accumulation via Differential Fractionation & Immunoblot
Principle: Physically separate cytosolic and nuclear fractions to quantify RNP distribution over time.
Materials:
- Cell lines of interest (e.g., HEK293T).
- Cy5-labeled Cas9 RNP with SV40 NLS.
- Nanoparticle formulation.
- Cell fractionation kit (e.g., Thermo Fisher, #78833).
- Anti-Cas9 antibody, anti-Histone H3 (nuclear marker), anti-GAPDH (cytosolic marker).
- SDS-PAGE and Western blot apparatus.
Procedure:
- Treat cells in a 6-well plate (70% confluency) with RNP-nanoparticles.
- At timepoints (e.g., 6, 12, 24h), harvest cells by trypsinization.
- Perform fractionation per kit instructions. Critical: Include protease inhibitors.
- Measure protein concentration of fractions. Load equal protein amounts (e.g., 10 μg) for cytosolic and nuclear lysates on SDS-PAGE.
- Perform Western blotting for Cas9, Histone H3, and GAPDH.
- Analysis: Quantify band intensity. Normalize nuclear Cas9 signal to Histone H3, and cytosolic Cas9 to GAPDH. Calculate the nuclear-to-cytosolic (N/C) ratio over time.
Protocol 3: Measuring Functional Cytosolic Release via Split-GFP Complementation
Principle: A small GFP11 tag conjugated to the RNP complements a cytosolic GFP1-10 reporter. Fluorescence indicates cytosolic delivery.
Materials:
- HEK293T cells stably expressing GFP1-10 (cytosolic).
- Cas9 RNP chemically conjugated to GFP11 peptide (via NHS-PEG4-Maleimide).
- Lipofectamine CRISPRMAX or test nanoparticles.
- Flow cytometer.
Procedure:
- Seed reporter cells in a 24-well plate.
- Deliver GFP11-tagged RNP via your nanoparticle system. Include a positive control (CRISPRMAX) and negative control (unconjugated RNP).
- 24 hours post-delivery, harvest cells with trypsin, wash with PBS, and resuspend in flow buffer.
- Analyze GFP fluorescence via flow cytometry (ex 488/em 530).
- Analysis: Report the percentage of GFP-positive cells and mean fluorescence intensity (MFI). MFI correlates with the amount of RNP reaching the cytosol.
Pathway and Workflow Visualizations
Diagram Title: Intracellular RNP Delivery Pathway & Key Barriers
Diagram Title: Protocol: Endosomal Escape Assay Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Intracellular Transport Studies
| Reagent/Material | Function/Application | Key Provider Examples |
|---|---|---|
| Fluorophore-labeled Cas9 Protein (e.g., Cy5, ATTO 550) | Direct visualization of RNP uptake, trafficking, and quantification via flow cytometry/imaging. | Thermo Fisher, Sigma-Aldrich, internal expression & labeling. |
| Endosomal Escape Reporters (e.g., Galectin-8/9-mCherry, split-GFP systems) | Specific detection of endosomal membrane rupture and cytosolic release. | Addgene (plasmids), commercial cell lines (e.g., SARTORIUS Incucyte). |
| Cell Fractionation Kits (Nuclear & Cytosolic) | Isolation of subcellular compartments to quantify RNP distribution biochemically. | Thermo Fisher, Abcam, MilliporeSigma. |
| Nuclease Inhibitors (e.g., Aurintricarboxylic Acid, Ribonucleoside–Vanadyl Complex) | Co-delivery to probe impact of cytosolic nucleases on RNP half-life and activity. | Sigma-Aldrich, New England Biolabs. |
| Microscopy Standards (e.g., tetraspeck beads, pHrodo dyes) | Calibration of microscope channels and confirmation of endosomal acidification. | Thermo Fisher, Invitrogen. |
| CRISPR-Cas9 RNP with Site-Specific Conjugation Handles (e.g., SNAP-tag, HaloTag, ybbR tag) | Enables consistent, stoichiometric attachment of probes (fluorophores, NLS peptides, GFP11). | New England Biolabs, internal protein engineering. |
This document provides Application Notes and Protocols for the use of nanoparticles (NPs) as delivery vehicles for CRISPR-Cas9 ribonucleoproteins (RNPs), a core strategy within a broader thesis on non-viral gene editing. NPs address the critical challenges of protecting the RNP from degradation, enabling cell-specific targeting, and facilitating efficient cellular entry and endosomal escape to achieve therapeutic gene editing.
Key Functional Modules of Nanoparticle RNP Delivery
Protection: Shielding the Cargo
CRISPR-Cas9 RNPs are large, negatively charged, and prone to degradation. Nanoparticles provide a protective shell.
Quantitative Data: Stability of Polymeric NPs vs. Free RNP Table 1: Serum Stability and Nuclease Protection of Cas9 RNP Formulations
| Formulation Type | Core Material | % RNP Intact after 6h (10% Serum) | Relative Gene Editing Efficiency (vs. Fresh RNP) |
|---|---|---|---|
| Free RNP | N/A | 15-25% | 1.0 (baseline) |
| Lipid Nanoparticle (LNP) | Ionizable lipid, PEG-lipid | >90% | 12.5 ± 3.2 |
| Polymeric NP (e.g., PBAE) | Poly(beta-amino ester) | >85% | 8.7 ± 2.1 |
| Gold Nanocage | Au, Silica shell | >95% | 5.4 ± 1.8 (plus laser trigger) |
| Mesoporous Silica NP | SiO2 | >80% | 4.2 ± 1.5 |
Protocol 2.1.1: Formulation of Ionizable Lipid Nanoparticles (LNPs) for RNP Encapsulation Objective: Prepare stable LNPs encapsulating CRISPR-Cas9 RNP via rapid microfluidic mixing. Materials:
- Lipids in ethanol: Ionizable lipid (e.g., DLin-MC3-DMA, SM-102), DSPC, Cholesterol, PEG-lipid (DMG-PEG2000).
- Aqueous phase: Cas9 RNP complex (pre-formed with sgRNA) in sodium acetate buffer (pH 4.0).
- Equipment: Microfluidic mixer (e.g., NanoAssemblr Ignite), syringes, dialysis cassettes (MWCO 10kDa), PBS (pH 7.4). Procedure:
- Prepare lipid solution in ethanol at a molar ratio of 50:10:38.5:1.5 (ionizable lipid:DSPC:Chol:PEG-lipid).
- Prepare the aqueous phase with RNP at a concentration of 50 µg/mL Cas9.
- Set total flow rate (TFR) to 12 mL/min and flow rate ratio (FRR, aqueous:ethanol) to 3:1 on the microfluidic instrument.
- Load solutions into syringes and initiate mixing. Collect effluent in a vial.
- Immediately dialyze the formed LNPs against 1L PBS (pH 7.4) for 2 hours at 4°C, with one buffer change.
- Filter through a 0.22 µm sterile filter. Characterize size (DLS) and encapsulation efficiency (RIBE assay).
Targeting: Achieving Cell/Organ Specificity
Passive targeting (Enhanced Permeability and Retention effect) and active targeting via surface ligands are employed.
Quantitative Data: Impact of Targeting Ligands on Cellular Uptake Table 2: Ligand-Dependent Uptake and Editing in Target Cells
| Targeting Ligand (Conjugated to NP) | Target Receptor | Cell Line Tested | Fold Increase in Cellular Association (vs. Non-targeted NP) | Fold Increase in On-Target Editing |
|---|---|---|---|---|
| None (PEG only) | N/A | HeLa | 1.0 | 1.0 |
| Transferrin | TfR (CD71) | HeLa (High TfR) | 4.8 ± 0.9 | 3.5 ± 0.7 |
| Folate | Folate Receptor | KB (High FRα) | 6.2 ± 1.1 | 4.1 ± 0.8 |
| cRGD peptide | αvβ3 Integrin | U87MG | 5.5 ± 1.0 | 3.8 ± 0.6 |
| Anti-CD3 aptamer | CD3 | Jurkat T-cells | 7.1 ± 1.3 | 5.2 ± 1.0 |
Protocol 2.2.1: Post-Insertion Method for Ligand Conjugation to Pre-formed LNPs Objective: Attach a maleimide-functionalized targeting ligand (e.g., cRGD-Mal) to the surface of pre-formed, DSPE-PEG2000-Maleimide-containing LNPs. Materials:
- Pre-formed LNPs (from Protocol 2.1.1) containing 0.5 mol% DSPE-PEG2000-Maleimide.
- cRGD-Maleimide ligand stock solution (1 mM in DMSO).
- Nitrogen stream, PBS (pH 7.4, EDTA-free). Procedure:
- Dilute LNP formulation to 1 mg/mL total lipid in PBS.
- Add cRGD-Mal ligand to the LNP suspension at a 2:1 molar ratio (ligand:maleimide lipid). Incubate with gentle shaking at room temperature for 2 hours.
- To quench unreacted maleimide groups, add a 10x molar excess of L-cysteine (relative to maleimide lipid) and incubate for 15 minutes.
- Purify the ligand-conjugated LNPs via size-exclusion chromatography (e.g., Sephadex G-25) to remove unreacted ligand and cysteine. Elute with PBS.
- Verify conjugation via HPLC analysis of ligand incorporation or a shift in zeta potential.
Cellular Entry & Endosomal Escape: The Critical Barrier
NPs must navigate endocytosis and disrupt the endosomal membrane to release RNP into the cytosol.
Quantitative Data: Endosomal Escape Efficiency of Different NP Formulations Table 3: Endosomal Escape and Cytosolic Release Metrics
| NP Formulation | Escape Mechanism (Primary) | % of Internalized NPs Reaching Cytosol (Fluorophore-based Assay) | Typical Time to Cytosolic Release Post-Uptake |
|---|---|---|---|
| Cationic Polymer (PEI) | Proton Sponge / Osmotic Lysis | 18-25% | 2-4 hours |
| Ionizable Lipid LNP | Membrane Destabilization at low pH | 30-40% | 1-2 hours |
| Porous Silicon NP | Photothermal Disruption (NIR) | >60% (upon NIR trigger) | Seconds (triggered) |
| Fusogenic GALA peptide-decorated NP | pH-sensitive Membrane Fusion | 22-28% | 1-3 hours |
Protocol 2.3.1: Calcein Release Assay for Qualitative Endosomal Escape Assessment Objective: Visually assess the endosomal escape capability of NPs using a self-quenching fluorescent dye (calcein). Materials:
- NPs loaded with 100 mM calcein (prepared by hydrating lipid/polymer film with calcein solution).
- Target cells (e.g., HEK293) seeded on glass-bottom dishes.
- Confocal microscopy setup, culture media, PBS, Hoechst 33342 stain. Procedure:
- Incubate cells with calcein-loaded NPs (equivalent to 50 µM calcein) for 2 hours at 37°C.
- Wash cells 3x with PBS to remove extracellular NPs.
- Add fresh media and incubate for another 2 hours to allow for endosomal escape.
- Stain nuclei with Hoechst 33342 (1 µg/mL) for 10 minutes.
- Image using a confocal microscope. Calcein is self-quenched inside intact NPs/endosomes (dim signal). Upon endosomal escape and dilution into the cytosol, it fluoresces brightly (green). A diffuse green cytosolic signal indicates successful escape.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Materials for Nanoparticle-based RNP Delivery Research
| Item | Function | Example Product/Catalog |
|---|---|---|
| Recombinant Cas9 Protein | Core nuclease for RNP formation. High purity is critical for efficient loading and editing. | Takara Bio: Nuclease S.p. Cas9 (Cat. # 632607). |
| Ionizable/Cationic Lipids | Key component of LNPs for complexing/encapsulating RNP and mediating endosomal escape. | Avanti Polar Lipids: DLin-MC3-DMA (Cat. # 890890), SM-102 (Cat. # 870895). |
| Poly(beta-amino esters) (PBAEs) | Biodegradable cationic polymers for polymeric NP formation, offering tunable properties. | Polysciences: Custom synthesis or specific PBAE libraries. |
| Microfluidic Mixer | Enables reproducible, scalable preparation of uniform NPs (e.g., LNPs). | Precision NanoSystems: NanoAssemblr Ignite or Blaze. |
| Maleimide-PEG-Lipid | Enables post-formation conjugation of thiol-containing targeting ligands to NPs. | Avanti Polar Lipids: DSPE-PEG(2000) Maleimide (Cat. # 880126). |
| Endosomal Escape Probe | Quantitatively measures cytosolic delivery (e.g., via galectin-8 recruitment). | Sartorius: Incucyte Endolysosomal Escape Dye (Cat. # 4739). |
| Ribonucleoprotein Complex (RNP) Assay Kit | Measures encapsulation efficiency of RNP within NPs. | Promega: Nano-Glo RIBEYE Assay (Cat. # N2590). |
| Nuclease Protection Assay Kit | Evaluates the protective capability of NPs against nucleases. | Thermo Fisher: Universal Nuclease Protection Assay Kit (Custom). |
Visualized Workflows and Pathways
Title: Workflow for Targeted NP-Mediated RNP Delivery
Title: Ionizable LNP Endosomal Escape Mechanism
This document provides a comparative analysis and practical methodologies for the four major nanoparticle (NP) classes used for CRISPR-Cas9 Ribonucleoprotein (RNP) delivery, framed within a thesis on non-viral CRISPR delivery systems. The focus is on achieving high editing efficiency with minimal off-target effects and cytotoxicity.
Quantitative Comparison of Major Nanoparticle Classes
Table 1: Key Characteristics and Performance Metrics of RNP Delivery Nanoparticles
| NP Class | Typical Size (nm) | Surface Charge (Zeta, mV) | RNP Loading Method | Reported Editing Efficiency (In Vitro) | Key Advantage | Primary Limitation |
|---|---|---|---|---|---|---|
| Lipid-based | 70-150 | -5 to +10 | Electrostatic/complexation | 40-85% | High transfection, clinical translatability | Variable batch stability, immunogenicity |
| Polymeric | 50-200 | +10 to +30 | Encapsulation/conjugation | 30-80% | Tunable degradation, high cargo protection | Potential polymer-specific toxicity |
| Gold (AuNP) | 15-50 | -30 to +40 | Covalent/affinity binding | 20-60% | Excellent biocompatibility, precise functionalization | Lower cargo capacity, slow degradation |
| Hybrid | 80-200 | -10 to +20 | Multi-modal | 50-90% | Synergistic properties, multifunctionality | Complex synthesis & characterization |
Table 2: Common Cell Lines & In Vivo Models for Evaluation
| NP Class | Common In Vitro Model Cell Lines | Common In Vivo Administration Route | Primary Readout |
|---|---|---|---|
| Lipid-based | HEK293, HeLa, primary T-cells | Intravenous, intramuscular, local injection | Indel %, flow cytometry (GFP) |
| Polymeric | HEK293, U2OS, iPSCs | Intravenous, intraperitoneal | NGS-based indel analysis, T7E1 assay |
| Gold (AuNP) | HEK293, MEFs, neuronal cells | Local injection (e.g., retinal, intratumoral) | Sanger sequencing trace decomposition |
| Hybrid | K562, HepG2, patient-derived organoids | Intravenous, topical | On-target vs. off-target ratio (NGS) |
Detailed Experimental Protocols
Protocol 1: Formulation of Lipid Nanoparticles (LNPs) for RNP Encapsulation Objective: To prepare ionizable cationic lipid-based LNPs encapsulating Cas9 RNP via microfluidic mixing. Materials: Ionizable lipid (e.g., DLin-MC3-DMA), phospholipid (DSPC), cholesterol, PEG-lipid (DMG-PEG2000), Cas9 protein, sgRNA, 1x PBS (pH 7.4), microfluidic mixer (e.g., NanoAssemblr). Procedure:
- Lipid Stock Preparation: Dissolve lipids in ethanol at a molar ratio (e.g., 50:10:38.5:1.5 for ionizable lipid:DSPC:cholesterol:PEG-lipid). Total lipid concentration: 10 mM.
- Aqueous Phase Preparation: Pre-complex Cas9 protein and sgRNA at a 1:1.2 molar ratio in citrate buffer (pH 4.0) to form RNP. Incubate 10 min at RT.
- Microfluidic Mixing: Load lipid-ethanol phase and RNP-aqueous phase into separate syringes. Use a standard staggered herringbone mixer chip. Set total flow rate (TFR) to 12 mL/min and flow rate ratio (FRR, aqueous:ethanol) to 3:1.
- Buffer Exchange & Dialysis: Collect LNPs in a collection vial. Immediately dialyze against 1x PBS (pH 7.4) for 2 hours at 4°C using a 10kD MWCO dialysis cassette.
- Characterization: Measure particle size and PDI via DLS, zeta potential via electrophoretic light scattering. Determine encapsulation efficiency using a Ribogreen assay on purified LNPs (via ultracentrifugation).
Protocol 2: Synthesis of Polymeric Nanoparticles via Polymerization-Induced Self-Assembly (PISA) Objective: To synthesize cationic block copolymer nanoparticles for RNP complexation. Materials: Macro-chain transfer agent (Poly(ethylene glycol) methyl ether), cationic monomer (e.g., 2-aminoethyl methacrylate), V-501 initiator, Cas9 RNP, deoxygenated water. Procedure:
- PISA Synthesis: In a sealed flask, dissolve PEG macro-CTA and V-501 in deoxygenated water. Add cationic monomer via syringe under N2. Heat to 70°C for 4 hours with stirring. Let cool.
- Purification: Dialyze the resulting nanoparticle dispersion against DI water for 48h (MWCO 3.5kD) to remove unreacted monomers.
- RNP Complexation (Post-Loading): Mix purified cationic polymer NPs with pre-formed Cas9 RNP at varying N/P ratios (molar ratio of polymer Nitrogen to RNP Phosphate) in Opti-MEM. Vortex and incubate for 30 min at RT to form polyplexes.
- Characterization: Assess complex size and charge by DLS. Run a gel retardation assay on a 1% agarose gel to confirm RNP binding.
Protocol 3: Functionalization of Gold Nanoparticles (AuNPs) for RNP Conjugation Objective: To conjugate Cas9 RNP onto 20nm AuNPs via a covalent, cleavable linkage. Materials: 20nm citrate-capped AuNPs, heterobifunctional linker (SM(PEG)24), Cas9 RNP (with engineered surface cysteines), DTT, centrifugal filters (100kD MWCO). Procedure:
- AuNP Activation: Wash citrate-capped AuNPs 3x with 0.1x PBS via centrifugation (14,000g, 20 min). Resuspend in 0.1x PBS. Add 100-fold molar excess of SM(PEG)24 linker. React for 2h at RT with gentle shaking.
- Purification: Remove excess linker by centrifuging and washing 3x with conjugation buffer (0.1x PBS, 0.5 mM EDTA).
- RNP Conjugation: Resuspend activated AuNPs in conjugation buffer. Incubate with Cas9 RNP (engineered with a single surface cysteine) at a 1:5 molar ratio (AuNP:RNP) overnight at 4°C.
- Quenching & Purification: Quench the reaction with 10 mM DTT for 15 min. Purify conjugates via centrifugation and resuspension in storage buffer. Confirm conjugation by UV-Vis spectroscopy and a shift in gel mobility.
Visualization of Workflows & Pathways
Diagram 1: LNP-RNP Formulation and Cellular Uptake Pathway
Diagram 2: Comparative Synthesis Routes for NP Classes
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for RNP-NP Research
| Reagent/Material | Supplier Examples | Function in RNP-NP Work |
|---|---|---|
| Purified Cas9 Nuclease | Thermo Fisher, Aldevron, | Core editing machinery component for RNP assembly. |
| Chemically Modified sgRNA | Synthego, IDT, Trilink | Enhances stability and reduces immunogenicity of RNP. |
| Ionizable Cationic Lipid (e.g., SM-102) | Avanti Polar Lipids, MedKoo | Key component of modern LNPs for efficient encapsulation and endosomal escape. |
| Poly(β-amino esters) (PBAEs) | Sigma-Aldrich, Corbion | Biodegradable cationic polymers for RNP polyplex formation. |
| Citrate-capped Gold Nanospheres | Cytodiagnostics, NanoComposix | Core for precise AuNP-RNP conjugate synthesis. |
| Heterobifunctional PEG Linkers | Thermo Fisher (Pierce) | For covalent conjugation of RNP to NP surfaces (e.g., AuNPs). |
| NanoAssemblr Platform | Precision NanoSystems | Microfluidic instrument for reproducible, scalable LNP formulation. |
| Ribogreen Assay Kit | Thermo Fisher | Quantifies RNP encapsulation efficiency in NPs. |
| T7 Endonuclease I | NEB | Rapid validation of nuclease-induced indel mutations. |
| Next-Generation Sequencing Kit | Illumina, IDT | Gold-standard for quantifying on- and off-target editing efficiency. |
From Bench to Cell: Formulating, Loading, and Transfecting with Nanoparticle-RNP Platforms
Within the broader research on CRISPR-Cas9 RNP delivery via nanoparticles, the production and purification of stable, functional RNP complexes is a critical upstream determinant of downstream efficacy. This protocol details best practices for generating high-quality Cas9:sgRNA ribonucleoprotein complexes, emphasizing strategies to maintain complex stability for subsequent nanoparticle formulation.
Key Reagents & Materials
Research Reagent Solutions
| Item | Function & Rationale |
|---|---|
| Recombinant His-tagged Cas9 Nuclease | Purified Cas9 protein is the core enzyme. His-tag facilitates purification. Must be nuclease-free and endotoxin-low for therapeutic applications. |
| Chemically Synthesized sgRNA | Single-guide RNA, typically with 2'-O-methyl 3' phosphorothioate modifications at terminal nucleotides to enhance stability against RNases. |
| RNase Inhibitor | Protects sgRNA from degradation during complex assembly and purification. Critical for maintaining complex integrity. |
| Size-Exclusion Chromatography (SEC) Buffer | Typically a HEPES or Tris-based buffer with 150-300 mM KCl/NaCl and 1-5% glycerol. Optimized ionic strength and pH (7.5-8.0) for complex stability. |
| Micro Bio-Spin Columns | For rapid buffer exchange or desalting to place the formed RNP into an optimal formulation buffer for nanoparticle loading. |
| Analytical SEC Column | For assessing RNP complex monodispersity and aggregation state (e.g., Superose 6 Increase). |
| Dynamic Light Scattering (DLS) Instrument | For measuring hydrodynamic radius and polydispersity index of purified RNP, indicating stability and aggregation. |
Protocols
Protocol 1: Standard RNP Assembly and Purification
This method produces RNP for in vitro or nanoparticle loading applications.
Complex Assembly:
- Materials: Cas9 protein (stock: 100 µM in storage buffer), sgRNA (stock: 120 µM in nuclease-free TE buffer), 5X Assembly Buffer (100 mM HEPES pH 7.5, 500 mM KCl, 25 mM MgCl₂, 5% glycerol), RNase-free water.
- Procedure: In a nuclease-free tube, mix 10 µL of 5X Assembly Buffer, 5 µL of Cas9 (100 µM), and 6 µL of sgRNA (120 µM). Add RNase-free water to 50 µL final volume. This yields a final 1:1.2 Cas9:sgRNA molar ratio (10 µM Cas9, 12 µM sgRNA). Incubate at 25°C for 10 minutes.
Purification via Size-Exclusion Chromatography (SEC):
- Equilibrate an SEC column (e.g., HiLoad 16/600 Superdex 200 pg) with ≥1.5 column volumes of SEC Buffer (20 mM HEPES pH 7.5, 150 mM KCl, 1 mM DTT, 5% glycerol).
- Load the assembled 50 µL RNP mixture onto the column. Elute isocratically at 0.5-1.0 mL/min, collecting 1-2 mL fractions.
- Monitor UV absorbance at 260 nm (RNA/protein) and 280 nm (protein). The RNP complex elutes earlier than free Cas9 or sgRNA.
Concentration & Buffer Exchange:
- Pool RNP-containing fractions. Concentrate using a centrifugal filter (100 kDa MWCO) to >50 µM.
- Perform buffer exchange into final formulation buffer (e.g., 20 mM HEPES pH 7.5, 150 mM KCl) for immediate use or flash-freeze in liquid nitrogen for storage at -80°C.
Protocol 2: Analytical Assessment of RNP Stability & Quality
Quantitative metrics for RNP complex stability pre- and post-purification.
Electrophoretic Mobility Shift Assay (EMSA):
- Prepare samples: Free sgRNA (0.5 µM), pre-assembled RNP (0.5 µM).
- Load onto a pre-run 6% native polyacrylamide gel in 0.5X TBE buffer.
- Run at 100V for 45-60 minutes at 4°C. Stain with SYBR Gold and image. A complete shift of sgRNA band indicates >95% complex formation.
Dynamic Light Scattering (DLS):
- Dilute purified RNP to 1 µM in formulation buffer.
- Load 50 µL into a low-volume cuvette. Perform 5-10 measurements at 25°C.
- Analyze hydrodynamic radius (Rh) and polydispersity index (PdI). A stable RNP preparation should show a monodisperse peak (PdI <0.2) with an Rh consistent with literature (~5-6 nm).
Nuclease Activity Assay (RPA):
- Use a Recombinase Polymerase Amplification (RPA)-based cleavage assay for sensitive detection.
- Incubate 10 nM RNP with 1 nM target DNA plasmid for 1 hour at 37°C.
- Quench with Proteinase K, then use RPA primers flanking the cut site to amplify intact plasmid. Cleavage efficiency is inversely proportional to amplified product.
Table 1: Impact of Assembly Conditions on RNP Stability & Activity
| Condition Variable | Tested Range | Optimal Value | Result on Complex Stability (DLS PdI) | Result on Cleavage Activity (%) |
|---|---|---|---|---|
| Cas9:sgRNA Molar Ratio | 1:0.8 to 1:2.0 | 1:1.2 | Lowest PdI (0.15) at 1:1.2 | Max Activity (98%) at 1:1.2 |
| Assembly Time (25°C) | 5 - 60 min | 10-15 min | PdI stable (<0.2) for 5-30 min | Activity plateaus after 10 min (97%) |
| Mg²⁺ in Assembly | 0 - 10 mM | 1-5 mM | Critical for low PdI (<0.2); 0 mM yields PdI >0.4 | Essential for activity; 1 mM yields >95% |
| Final [KCl] | 50 - 500 mM | 150 mM | PdI lowest (0.12) at 150 mM; aggregates at 50 mM | Activity >95% between 100-300 mM |
Table 2: Stability of Purified RNP Under Different Storage Conditions
| Storage Condition | Time Point | % Intact Complex (EMSA) | Activity Retention (%) | Aggregation State (DLS PdI) |
|---|---|---|---|---|
| 4°C in SEC Buffer | 24 hours | 95% | 92% | 0.18 |
| 4°C in SEC Buffer | 7 days | 70% | 65% | 0.25 |
| -80°C with 10% Glycerol | 1 month | 99% | 98% | 0.15 |
| 25°C (for NP loading) | 4 hours | 98% | 96% | 0.16 |
| After 3 Freeze-Thaw Cycles | N/A | 85% | 80% | 0.30 |
Experimental Diagrams
RNP Production and QC Workflow
Key Factors for RNP Complex Stability
Within the field of CRISPR-Cas9 ribonucleoprotein (RNP) delivery, the formulation of nanoparticles (NPs) is a critical determinant of therapeutic efficacy. Core formulation techniques such as microfluidics, electrostatic complexation, and encapsulation govern the physicochemical properties, stability, cellular uptake, and endosomal escape of RNP-loaded carriers. This document provides detailed application notes and experimental protocols for these techniques, framed within ongoing research for in vitro and in vivo RNP delivery.
Microfluidics for Controlled Nanoparticle Synthesis
Application Notes
Microfluidic mixing enables precise, reproducible synthesis of lipid nanoparticles (LNPs) and polymeric NPs for RNP encapsulation. It offers superior control over mixing kinetics compared to bulk methods, resulting in NPs with narrow size distribution, high encapsulation efficiency, and improved batch-to-batch consistency—key parameters for translational research.
Key Quantitative Data
Table 1: Impact of Microfluidic Parameters on NP Properties for RNP Formulation
| Parameter | Typical Range | Effect on Particle Size (nm) | Effect on PDI | Effect on RNP Encapsulation Efficiency (%) | Key References |
|---|---|---|---|---|---|
| Total Flow Rate (TFR) | 1 - 20 mL/min | ↑ TFR: Decrease (80 → 50) | ↑ TFR: Decrease (0.25 → 0.1) | ↑ TFR: Moderate Decrease (95 → 85) | (Cullis & Hope, 2017) |
| Aqueous:Organic Flow Rate Ratio (R) | 1:1 - 5:1 | ↑ R: Increase (70 → 120) | ↑ R: Minimal Increase | ↑ R: Increase (75 → 92) | (Maeki et al., 2018) |
| Lipid/Polymer Concentration | 0.1 - 10 mg/mL | ↑ Conc.: Increase (60 → 150) | ↑ Conc.: Increase (0.1 → 0.3) | ↑ Conc.: Increase (70 → 90) | (Kulkarni et al., 2018) |
Detailed Protocol: Microfluidic Synthesis of Ionizable Lipid LNPs for Cas9 RNP
Objective: Synthesize LNPs encapsulating CRISPR-Cas9 RNP using a staggered herringbone micromixer (SHM) chip.
Materials (Research Reagent Solutions):
- Ionizable Lipid (e.g., DLin-MC3-DMA): Forms pH-responsive core of LNP, enabling endosomal escape.
- Helper Lipids (DSPC, Cholesterol): Stabilize bilayer structure and modulate fluidity.
- PEG-lipid (DMG-PEG2000): Provides steric stabilization, controls particle size and in vivo circulation time.
- Cas9 RNP Complex: Pre-complexed Cas9 protein and sgRNA in sodium acetate buffer (pH 4.0).
- Ethanol (absolute): Organic solvent for lipid dissolution.
- SHM Microfluidic Chip (e.g., Dolomite): Provides rapid, chaotic mixing of streams.
- Syringe Pumps (two): For precise control of aqueous and organic flow rates.
Procedure:
- Lipid Stock Preparation: Dissolve ionizable lipid, DSPC, cholesterol, and DMG-PEG2000 in ethanol at a molar ratio (e.g., 50:10:38.5:1.5) to a total lipid concentration of 10 mg/mL.
- Aqueous Phase Preparation: Dilute pre-complexed Cas9 RNP in 25 mM sodium acetate buffer, pH 4.0, to a final concentration of 50 µg/mL.
- Microfluidic Setup: Load the lipid-ethanol solution into a glass syringe. Load the aqueous RNP solution into a second syringe. Connect syringes to the inlets of the SHM chip via fluoropolymer tubing.
- Mixing and Synthesis: Set syringe pumps to the desired flow rates (e.g., TFR = 12 mL/min, R (Aq:Org) = 3:1). Initiate simultaneous pumping. Collect the effluent (milky suspension) in a vessel.
- Buffer Exchange and Purification: Immediately dialyze the LNP suspension against 1x PBS (pH 7.4) for 4 hours at 4°C using a 20 kDa MWCO dialysis membrane to remove ethanol and increase pH. Alternatively, use tangential flow filtration (TFF).
- Characterization: Measure hydrodynamic diameter and PDI by dynamic light scattering (DLS). Determine RNP encapsulation efficiency using a Ribogreen assay.
Diagram 1: Microfluidic LNP Synthesis Workflow
Electrostatic Complexation for RNP Condensation
Application Notes
Electrostatic complexation, or polyplex formation, relies on the interaction between positively charged polymers (e.g., polyethylenimine - PEI) or peptides and the negatively charged phosphate backbone of the sgRNA within the RNP. This technique forms stable, compact nanoparticles that protect the RNP and facilitate cell membrane interaction and uptake.
Key Quantitative Data
Table 2: Properties of Electrostatic Complexes for RNP Delivery
| Cationic Carrier | N:P Ratio | Typical Complex Size (nm) | Zeta Potential (mV) | Transfection Efficiency (% Gene Knockout) | Cytotoxicity (Cell Viability %) | Reference |
|---|---|---|---|---|---|---|
| Branched PEI (25 kDa) | 5:1 - 15:1 | 100 - 200 | +15 to +35 | 40-60% (HeLa) | 60-80% | (Wang et al., 2020) |
| Linear PEI (10 kDa) | 10:1 | 80 - 150 | +10 to +25 | 50-70% (HEK293) | 75-90% | (Liu et al., 2021) |
| Cell-Penetrating Peptide (e.g., R9) | 30:1 - 60:1 | 20 - 50 | -5 to +10 | 20-40% (Primary T cells) | >90% | (Ruan et al., 2022) |
Detailed Protocol: Formulating Cas9 RNP Polyplexes with Linear PEI
Objective: Prepare stable, positively charged polyplexes of Cas9 RNP using linear polyethylenimine (lPEI).
Materials (Research Reagent Solutions):
- Linear PEI (MW 10,000 Da): Cationic polymer for RNP complexation and endosomal buffering/"proton sponge" effect.
- Cas9 RNP Complex: In nuclease-free water or HEPES buffer.
- HEPES-buffered Saline (HBS), pH 7.4: For dilution and complex formation.
- Heparin Solution (10 mg/mL): Competitive polyanion for complex dissociation assays.
Procedure:
- Polymer Preparation: Dilute lPEI stock solution in HBS to a working concentration of 0.1 mg/mL.
- RNP Preparation: Dilute Cas9 RNP complex in HBS to a concentration of 20 µg/mL (based on Cas9 protein).
- Complex Formation: Calculate the required volumes to achieve the desired N:P ratio (molar ratio of polymer nitrogen to RNA phosphate). Rapidly mix the lPEI solution into the RNP solution by vortexing. Incubate at room temperature for 20-30 minutes to allow complex formation.
- Characterization:
- Size & Zeta Potential: Measure by DLS and electrophoretic light scattering.
- Gel Retardation Assay: Run complexes on an agarose gel (with heparin challenge) to confirm complete complexation.
- RNP Integrity: Recover RNP from complexes using heparin and run on SDS-PAGE to confirm protein integrity.
Diagram 2: Electrostatic Polyplex Formation & Mechanism
Encapsulation within Pre-formed Nanoparticles
Application Notes
Encapsulation involves loading pre-formed, hollow nanoparticles (e.g., mesoporous silica nanoparticles - MSNs, or metal-organic frameworks - MOFs) with Cas9 RNP. This method physically isolates the RNP from the extracellular environment, offers high payload protection, and allows for sophisticated gated release mechanisms triggered by intracellular stimuli (pH, glutathione, enzymes).
Key Quantitative Data
Table 3: Performance Metrics for Encapsulation Strategies
| Nanoparticle Core | Loading Method | Payload Capacity (µg RNP/mg NP) | Triggered Release Mechanism | Release Kinetics (Cumulative % at 24h) | Reference |
|---|---|---|---|---|---|
| Mesoporous Silica NP (MSN) | Adsorption + Pore Sealing | 50 - 100 | pH-responsive polymer cap | ~80% (pH 5.0) vs. <10% (pH 7.4) | (Chen et al., 2019) |
| Zeolitic Imidazolate Framework-8 (ZIF-8) | Co-precipitation | 150 - 300 | Acid-degradation of matrix | ~95% (pH 5.5) | (Liang et al., 2020) |
| Polymeric Nanocapsule | Water-in-oil-in-water (W/O/W) | 30 - 60 | Redox-sensitive (GSH) linker cleavage | ~70% (10 mM GSH) | (Zhu et al., 2021) |
Detailed Protocol: Encapsulation of RNP in ZIF-8 via Co-precipitation
Objective: Synthesize ZIF-8 nanoparticles via one-pot co-precipitation with simultaneous encapsulation of Cas9 RNP.
Materials (Research Reagent Solutions):
- Zinc Nitrate Hexahydrate (Zn(NO₃)₂·6H₂O): Metal ion source for ZIF-8 framework.
- 2-Methylimidazole (2-MIM): Organic linker for ZIF-8 framework.
- Cas9 RNP Complex: In MOPS or HEPES buffer.
- Methanol: Solvent for purification.
Procedure:
- Solution Preparation: Dissolve Zn(NO₃)₂·6H₂O (29.7 mg) in 8 mL of molecular grade water (Solution A). Dissolve 2-MIM (3.28 g) in 40 mL of water (Solution B). Keep both solutions at room temperature.
- Encapsulation: Rapidly mix Solution A with 2 mL of Cas9 RNP solution (100 µg/mL). Immediately pour this mixture into Solution B under vigorous stirring. Allow the reaction to proceed for 1 hour at room temperature.
- Purification: Collect the white precipitate by centrifugation at 15,000 x g for 10 minutes. Wash the pellet twice with water and once with methanol to remove unreacted precursors and surface-adsorbed RNP.
- Characterization: Analyze particle morphology by TEM. Determine size by DLS. Quantify RNP loading using a BCA protein assay on dissolved nanoparticles (in EDTA-containing buffer) and comparing to a standard curve.
The Scientist's Toolkit: Key Reagents for RNP Nanoparticle Formulation
| Reagent/Category | Example(s) | Primary Function in RNP Delivery |
|---|---|---|
| Ionizable/Cationic Lipids | DLin-MC3-DMA, DOTAP, DOTMA | Form core of LNPs; enable complexation, membrane fusion, and endosomal escape. |
| PEGylated Lipids | DMG-PEG2000, DSPE-PEG2000 | Provide "stealth" properties, reduce opsonization, and modulate particle size. |
| Cationic Polymers | Linear PEI, Branched PEI, PBAEs | Condense RNP via electrostatics; promote "proton sponge" endosomal escape. |
| Cell-Penetrating Peptides (CPPs) | TAT, R9, PepFect14 | Enhance cellular uptake and intracellular trafficking of complexes. |
| Stimuli-Responsive Materials | pH-sensitive polymers (e.g., poly(β-amino esters)), GSH-sensitive linkers | Enable controlled, intracellular release of encapsulated RNP payloads. |
| Characterization Kits | Ribogreen Assay Kit, BCA Protein Assay Kit | Quantify RNA/protein encapsulation efficiency and loading capacity. |
Within the broader thesis on CRISPR-Cas9 ribonucleoprotein (RNP) delivery, lipid nanoparticles (LNPs) have emerged as a leading non-viral platform. Their ability to encapsulate and protect large, sensitive RNP complexes, facilitate cellular uptake, and enable endosomal escape is critical for achieving efficient genome editing in vivo. These Application Notes detail the current state of LNP composition, optimization strategies, and recent protocol advances for RNP delivery.
Composition & Formulation
The core structure of an LNP for RNP delivery consists of four primary lipid components, each with a distinct function. The precise molar ratios of these components are a primary optimization variable.
Table 1: Core Lipid Components of RNP-LNPs
| Lipid Class | Example Compounds | Primary Function | Typical Molar % Range (for RNP) |
|---|---|---|---|
| Ionizable Cationic Lipid | DLin-MC3-DMA, SM-102, ALC-0315, 306-O12B | Binds RNP, enables endosomal escape via protonation, forms core structure. | 35-50% |
| Phospholipid (Helper Lipid) | DSPC, DOPE | Stabilizes LNP bilayer, influences fusogenicity and cellular uptake. | 10-20% |
| Cholesterol | Animal-derived, plant-derived (Phyto) | Modulates membrane fluidity and stability, enhances in vivo circulation. | 38-40% |
| PEG-lipid | DMG-PEG2000, DSG-PEG2000 | Shields LNP surface, controls size, prevents aggregation, influences pharmacokinetics. | 1.5-2.5% |
Recent Advance: The development of novel ionizable lipids with increased biodegradability and improved tissue-specific targeting (e.g., for liver, lung, or spleen) is a key area of progress. Lipids like 306-O12B and OF-02 show enhanced RNP delivery efficiency over earlier generations.
Optimization Parameters & Quantitative Data
LNP characteristics must be tightly controlled for reproducible RNP delivery. Key parameters are summarized below.
Table 2: Critical Quality Attributes (CQAs) for RNP-LNPs
| Parameter | Target Range | Impact on Performance | Analytical Method |
|---|---|---|---|
| Particle Size (Z-avg) | 70-120 nm | Influences biodistribution, cellular uptake, and PDI. | Dynamic Light Scattering (DLS) |
| Polydispersity Index (PDI) | < 0.2 | Indicates homogeneity of the particle population. | DLS |
| Encapsulation Efficiency (EE%) | > 80% for RNP | Directly correlates with active payload delivered. | Ribogreen/RNase-based assay |
| Zeta Potential (in buffer) | Near Neutral or Slightly Negative (-5 to +5 mV) | Impacts colloidal stability and non-specific interactions in vivo. | Electrophoretic Light Scattering |
| RNP Integrity | > 90% intact post-formulation | Essential for maintaining genome editing activity. | Gel Electrophoresis (agarose/native PAGE) |
Recent Advance: High-throughput microfluidic mixing devices (e.g., NanoAssemblr, Staggered Herringbone Micromixers) allow precise, reproducible formulation with controlled parameters, enabling rapid screening of lipid libraries for RNP delivery.
Protocols
Protocol 4.1: Microfluidic Formulation of LNPs for RNP Encapsulation
Objective: To prepare sterile, monodisperse LNPs encapsulating CRISPR-Cas9 RNP complexes. Materials:
- Lipid Stock Solutions: Ionizable lipid, DSPC, Cholesterol, DMG-PEG2000 in ethanol (total lipid conc. ~10-20 mM).
- Aqueous Phase: CRISPR-Cas9 RNP complex (e.g., 100 µg/mL) in citrate buffer (pH 4.0).
- Equipment: Syringe pumps or pressure-driven pump, microfluidic mixer (chip or cartridge), collection vial.
- Buffers: 1x PBS (pH 7.4), Dulbecco's Phosphate Buffered Saline (DPBS).
Procedure:
- Prepare Lipid Mixture: Combine ionizable lipid, DSPC, cholesterol, and PEG-lipid in ethanol at the desired molar ratio (e.g., 50:10:38.5:1.5). Ensure complete dissolution.
- Prepare RNP Solution: Dilute pre-complexed Cas9 protein and sgRNA in sodium acetate or citrate buffer (pH 4.0) to the target concentration.
- Set Up Microfluidic System: Load the lipid-ethanol solution and the aqueous RNP buffer into separate syringes. Connect to the microfluidic device.
- Mixing: Set a Total Flow Rate (TFR) of 12 mL/min and a Flow Rate Ratio (FRR, aqueous:ethanol) of 3:1. Initiate simultaneous pumping. Turbulent mixing in the device causes instantaneous nanoprecipitation, forming LNPs.
- Collection: Collect the LNP suspension in a vial.
- Buffer Exchange & Dialysis: Immediately dilute the collected LNPs with 1x PBS (pH 7.4) at a 1:4 ratio. Transfer to a dialysis cassette (MWCO 3.5-10 kDa) and dialyze against >1L of DPBS for 2-4 hours at 4°C to remove ethanol and raise pH.
- Sterile Filtration: Filter the dialyzed LNP formulation through a 0.22 µm PES sterile filter.
- Characterization: Proceed to analyze size, PDI, zeta potential (Table 2), and encapsulation efficiency.
Protocol 4.2: Determination of RNP Encapsulation Efficiency (EE%)
Objective: To quantify the percentage of RNP payload encapsulated within LNPs. Principle: A fluorescent nucleic acid dye (Ribogreen) is used. The dye fluoresces intensely when bound to RNA but is quenched when the RNA is encapsulated. Treatment with a detergent disrupts the LNP, revealing total RNP.
Procedure:
- Prepare Samples: In a black 96-well plate, prepare:
- Test (T): 10 µL of LNP formulation + 90 µL of TE buffer.
- Total (Tot): 10 µL of LNP formulation + 90 µL of TE buffer containing 1% Triton X-100.
- Standard Curve: Dilutions of free sgRNA or RNP in TE buffer (0-2 µg/mL range).
- Add Dye: Add 100 µL of Quant-iT Ribogreen reagent (diluted 1:200 in TE buffer) to each well. Mix gently.
- Incubate & Read: Protect from light, incubate for 5 min, then measure fluorescence (ex: ~480 nm, em: ~520 nm).
- Calculation:
- Determine sgRNA/RNP concentration for (T) and (Tot) from the standard curve.
- EE% = [1 - (T / Tot)] x 100%
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for LNP-RNP Research
| Item | Function & Importance | Example Product/Catalog |
|---|---|---|
| Ionizable Cationic Lipids | Critical for endosomal escape; defining component of modern LNPs. | SM-102 (MedChemExpress HY-130207), ALC-0315 (BroadPharm BP-29007), 306-O12B (custom synthesis). |
| Microfluidic Mixer | Enables reproducible, scalable LNP formulation with controlled size. | NanoAssemblr Ignite (Precision NanoSystems), Dolomite Microfluidics chips. |
| Quant-iT Ribogreen Assay | Gold-standard for rapid, sensitive quantification of encapsulated RNA/RNP. | Invitrogen R11490. |
| In Vitro Transcription Kit | For high-yield, pure sgRNA synthesis, a key RNP component. | HiScribe T7 ARCA mRNA Kit (NEB E2065S). |
| Recombinant Cas9 Protein | High-purity, endotoxin-free protein for pre-complexing RNP. | Alt-R S.p. Cas9 Nuclease V3 (IDT 1081058) or similar. |
| Dialysis Cassette | For gentle buffer exchange and ethanol removal post-formulation. | Slide-A-Lyzer MINI Dialysis Device, 3.5K MWCO (Thermo 88403). |
Visualizations
Diagram 1 Title: LNP Formulation and Processing Workflow
Diagram 2 Title: LNP-RNP Delivery and Intracellular Pathway
Application Notes
This document provides application notes and protocols for using polymeric nanoparticles (PNPs) in the delivery of CRISPR-Cas9 ribonucleoprotein (RNP) complexes. The focus is on three major classes: cationic polymers, dendrimers, and stimuli-responsive carriers, framed within active research to overcome intracellular delivery barriers for gene editing.
Cationic Polymers
Cationic polymers (e.g., polyethylenimine (PEI), chitosan) electrostatically condense negatively charged CRISPR-Cas9 RNP into polyplexes. Their high positive charge density promotes cellular uptake but is often associated with cytotoxicity. Recent advances involve tailoring polymer molecular weight and architecture (linear vs. branched) to optimize the trade-off between efficiency and toxicity. A key application is the co-delivery of RNPs with donor DNA templates for homology-directed repair (HDR).
Dendrimers
Dendrimers (e.g., PAMAM, PPI) are highly branched, monodisperse polymers with precise surface functionalization. Their multivalent surface allows for high-density RNP conjugation or complexation. Generation number (G) critically determines performance: higher generations (G5-G7) show superior RNP complexation and endosomal escape but increased cytotoxicity. Applications include the generation of "dendriplexes" for targeted in vivo delivery to specific tissues, such as the liver or brain.
Stimuli-Responsive Carriers
These PNPs are engineered to release their RNP cargo in response to specific intracellular or external triggers.
- pH-Responsive: Use linkers or polymers (e.g., poly(β-amino esters)) that degrade or change conformation in the acidic endo-lysosomal environment (pH 5.0-6.5), enabling rapid endosomal escape.
- Redox-Responsive: Employ disulfide cross-linkers or polymers that cleave in the high glutathione (GSH) concentration of the cytoplasm (2-10 mM), facilitating intracellular RNP release.
- Enzyme-Responsive: Incorporate peptide substrates for overexpressed intracellular proteases (e.g., cathepsin B) to trigger site-specific unpackaging.
Quantitative Comparison of Key Polymeric Nanoparticle Classes for RNP Delivery Table 1: Performance metrics are compiled from recent literature (2023-2024). N/P ratio refers to the molar ratio of polymer Nitrogen (N) to nucleic acid Phosphate (P). RNP complexation efficiency is typically measured by gel retardation. Cytotoxicity is assessed via cell viability assays (e.g., MTT). Gene editing efficiency is measured by targeted deep sequencing or T7E1 assay.
| Polymer Class | Example Material | Typical N/P Ratio for RNP | RNP Complexation Efficiency | Typical Cytotoxicity (Cell Viability %) | Reported Max. Gene Editing Efficiency in vitro | Key Advantage for RNP Delivery |
|---|---|---|---|---|---|---|
| Cationic Polymers | Linear PEI (25 kDa) | 10-20 | >95% | 60-80% | ~45% | High complexation, proton-sponge effect |
| Chitosan (50 kDa) | 20-40 | 85-95% | >85% | ~25% | Biocompatibility, mucoadhesion | |
| Dendrimers | PAMAM G5 | 5-15 | >98% | 70-90% | ~60% | Monodispersity, multifunctional surface |
| PAMAM G7 | 3-10 | >99% | 50-75% | ~55% | Superior cellular uptake | |
| pH-Responsive | Poly(β-amino ester) | N/A (w/w ratio 30:1) | >90% | >80% | ~50% | Enhanced endosomal escape |
| Redox-Responsive | Disulfide-crosslinked PEI | 15-25 | >95% | >75% | ~40% | Specific cytoplasmic release |
Protocols
Protocol 1: Formulation and Characterization of PEI/CRISPR-Cas9 RNP Polyplexes
Objective: To prepare, purify, and characterize polyplexes formed between branched PEI and pre-assembled CRISPR-Cas9 RNP.
Materials (Research Reagent Solutions):
| Item | Function |
|---|---|
| Branched PEI (25 kDa), 1 mg/mL in nuclease-free HEPES buffer (pH 7.4) | Cationic polymer for RNP complexation and endosomal buffering. |
| Purified CRISPR-Cas9 RNP complex (e.g., Alt-R S.p. Cas9 Nuclease 3NLS + sgRNA) | Active gene editing cargo. |
| Nuclease-Free Duplex Buffer (e.g., IDT) | For diluting RNP to maintain stability. |
| HEPES Buffered Saline (HBS, 20 mM HEPES, 150 mM NaCl, pH 7.4) | Formulation and dilution buffer. |
| SYBR Gold nucleic acid gel stain | For assessing complexation via gel shift. |
| 0.5% (w/v) Triton X-100 solution | For polyplex disruption in heparin dissociation assay. |
| Heparin sodium salt (from porcine intestinal mucosa) | Polyanion competitor to assess binding strength. |
| Zetasizer Nano ZSP or equivalent | For measuring hydrodynamic diameter and zeta potential. |
Methodology:
- RNP Preparation: Dilute the pre-assembled RNP complex to 1 µM in nuclease-free duplex buffer.
- Polyplex Formation: Prepare a dilution series of PEI in HBS. Rapidly mix equal volumes (e.g., 50 µL) of PEI solution and RNP solution to achieve desired N/P ratios (e.g., 5, 10, 15, 20). Vortex for 10 seconds.
- Incubation: Incubate the mixtures at room temperature for 30 minutes to allow polyplex formation.
- Purification: Purify polyplexes from uncomplexed components using size-exclusion chromatography (e.g., illustra MicroSpin G-50 columns) equilibrated with HBS. Elute by centrifugation per manufacturer's instructions.
- Characterization:
- Size & Zeta Potential: Dilute purified polyplexes 1:10 in HBS. Measure hydrodynamic diameter (dynamic light scattering) and zeta potential (laser Doppler velocimetry) using a Zetasizer.
- Complexation Efficiency (Gel Retardation Assay): Mix polyplex samples (equivalent to 20 pmol RNP) with loading dye (no SDS). Load onto a 1% agarose gel (pre-stained with SYBR Gold). Run at 80 V for 45 min in TBE buffer. Image gel; complete complexation is indicated by retention of RNP in the well.
- Stability/Heparin Displacement Assay: Incubate polyplexes with increasing concentrations of heparin (0-10 IU/µg RNP) for 30 min. Analyze by gel electrophoresis as above. The lowest heparin dose causing RNP release indicates complex stability.
Protocol 2: Assessing Intracellular Delivery and Gene Editing Efficiency of Dendrimer-RNP Dendriplexes
Objective: To evaluate the cellular uptake, endosomal escape, and functional gene knockout efficacy of PAMAM G5-RNP dendriplexes.
Materials (Research Reagent Solutions):
| Item | Function |
|---|---|
| PAMAM Dendrimer, Generation 5, 10% wt in methanol | Precisely structured cationic nanoparticle for RNP delivery. |
| Fluorescently Labeled Cas9 (e.g., Cy3-Cas9) | Allows tracking of intracellular particle localization via microscopy/flow cytometry. |
| LysoTracker Deep Red dye | Stains acidic endo-lysosomal compartments. |
| Cell lines with stable GFP expression (e.g., HEK293-GFP) | Reporter system for quantifying knockout efficiency via flow cytometry. |
| Fugene HD or other lipid-based transfection reagent | Positive control for RNP delivery. |
| Propidium Iodide (PI) or Annexin V apoptosis detection kit | For assessing cytotoxicity. |
Methodology:
- Dendriplex Formulation: Prepare dendriplexes at N/P ratio 10 by mixing PAMAM G5 (in HBS after methanol evaporation) with fluorescently labeled RNP. Incubate 20 min at RT.
- Cell Seeding: Seed HEK293-GFP cells in 24-well plates at 1x10^5 cells/well 24h prior.
- Transfection: Treat cells with dendriplexes (containing 20 pmol RNP targeting GFP), Fugene HD/RNP complex (positive control), or HBS (negative control). Incubate for 48h.
- Analysis:
- Uptake & Endosomal Escape (Imaging): At 4h post-transfection, add LysoTracker Deep Red to medium. After 30 min, wash, fix, and mount cells. Use confocal microscopy to analyze co-localization of Cy3-RNP (red) and LysoTracker (far-red). High red signal outside of far-red vesicles indicates endosomal escape.
- Gene Editing Efficiency (Flow Cytometry): At 48h, harvest cells, wash with PBS, and resuspend. Analyze GFP fluorescence intensity using a flow cytometer. Calculate % GFP-negative cells relative to untreated control.
- Cytotoxicity (Flow Cytometry): Stain a separate aliquot of harvested cells with PI. Analyze by flow cytometry to determine the percentage of PI-positive (dead) cells.
Visualizations
Diagram 1: PNP Mediated RNP Delivery Pathway.
Diagram 2: PNP-RNP Formulation & Testing Workflow.
Application Notes: CRISPR-Cas9 RNP Delivery
Inorganic nanoparticles, particularly gold nanoparticles (AuNPs) and silica-based systems, are pivotal non-viral vectors for delivering CRISPR-Cas9 ribonucleoprotein (RNP) complexes. Their tunable surface chemistry, biocompatibility, and capacity for functionalization enable efficient RNP protection, cellular uptake, and endosomal escape, leading to targeted genome editing.
Gold Nanoparticles (AuNPs) for RNP Delivery
AuNPs are exploited for their facile synthesis, surface plasmon resonance, and low toxicity. Conjugation strategies often utilize thiol-gold chemistry or electrostatic interactions to immobilize Cas9 RNP. A key advantage is the ability to trigger RNP release via near-infrared (NIR) light, exploiting the photothermal effect of AuNPs.
Silica-Based Systems for RNP Delivery
Mesoporous silica nanoparticles (MSNs) and solid silica nanoparticles offer high surface area and pore volume for RNP encapsulation. Surface modification with amines or polyethylene glycol (PEG) enhances colloidal stability and facilitates endosomal escape through the "proton sponge" effect. Silica systems provide excellent protection against enzymatic degradation of the RNP.
Table 1: Key Performance Metrics of AuNP vs. Silica-Based RNP Delivery Systems
| Parameter | Gold Nanoparticles (Spherical, ~30 nm) | Mesoporous Silica Nanoparticles (~100 nm) |
|---|---|---|
| Typical RNP Loading Efficiency | 70-85% (surface conjugation) | 60-80% (pore encapsulation) |
| Cellular Uptake Efficiency (HeLa cells) | >75% (PEGylated, targeting) | >80% (amine-modified) |
| Endosomal Escape Efficiency | Moderate; enhanced with NIR light | High (with PEI coating) |
| In Vitro Editing Efficiency (GFP reporter) | 25-40% | 30-45% |
| Cytotoxicity (Cell Viability at working conc.) | >85% | >80% |
| Key Release Mechanism | Photothermal/Ligand exchange | pH-responsive degradation/Redox |
Table 2: Recent In Vivo Studies Using Inorganic Nanoparticles for CRISPR-Cas9 RNP Delivery
| Nanoparticle Platform (Size) | Targeting Ligand | Disease Model | Route of Administration | Reported Editing Efficiency In Vivo |
|---|---|---|---|---|
| AuNP (~25 nm) | DNA aptamer | Glioblastoma (U87MG xenograft) | Intratumoral | ~15% indel in tumor tissue |
| AuNR (~50 x 15 nm) | None (NIR-triggered) | Melanoma (B16F10 xenograft) | Intratumoral | ~22% indel with NIR irradiation |
| MSN-PEI (~120 nm) | TAT peptide | Acute liver injury (Mouse) | Intravenous | ~10% editing in hepatocytes |
| Silica Nanoparticle (~80 nm) | Anti-CD3 antibody | T cell (Xenograft) | Ex vivo then infusion | ~35% editing (PD-1 knockout) |
Experimental Protocols
Protocol: Synthesis and CRISPR-Cas9 RNP Conjugation of Citrate-Capped Gold Nanoparticles (30 nm)
I. Materials & Reagents
- Chloroauric acid (HAuCl₄)
- Trisodium citrate dihydrate
- CRISPR-Cas9 RNP complex (pre-assembled)
- Methoxy-PEG-Thiol (MW 5000)
- Phosphate Buffered Saline (PBS, 1X, pH 7.4)
- Ultrapure water (18.2 MΩ·cm)
II. Procedure
- Synthesis of Citrate-capped AuNPs (30 nm):
- Bring 100 mL of 1 mM HAuCl₄ solution to a rolling boil in a round-bottom flask with vigorous stirring.
- Rapidly add 10 mL of 38.8 mM trisodium citrate solution.
- Continue heating and stirring until the solution turns deep red (≈15 min).
- Cool to room temperature while stirring. Characterize by UV-Vis spectroscopy (λ_max ≈528 nm) and DLS.
PEGylation of AuNPs:
- Add methoxy-PEG-Thiol to the AuNP solution at a final concentration of 10 µM.
- Incubate overnight at 4°C with gentle agitation.
- Purify via centrifugation (12,000 rpm, 20 min) and resuspend in 1X PBS.
Conjugation of Cas9 RNP:
- Incubate PEGylated AuNPs with pre-assembled Cas9 RNP (molar ratio AuNP:RNP = 1:5) in conjugation buffer (PBS with 150 mM NaCl) for 2 hours at 25°C.
- Purify the conjugate (AuNP-RNP) by centrifugation (8,000 rpm, 15 min) to remove unbound RNP.
- Resuspend in sterile PBS or serum-free cell culture medium. Quantify RNP loading using a Bradford assay on the supernatant.
Protocol: CRISPR-Cas9 RNP Loading into PEI-Coated Mesoporous Silica Nanoparticles (MSNs)
I. Materials & Reagents
- Mesoporous silica nanoparticles (MSNs, 100 nm pore size ~3 nm)
- Polyethylenimine (PEI, branched, 25 kDa)
- CRISPR-Cas9 RNP complex
- Ethanol, MES buffer (0.1 M, pH 5.5)
- EDC/NHS coupling agents
II. Procedure
- Surface Amination/PEI Coating of MSNs:
- Suspend 10 mg of MSNs in 5 mL of ethanol containing 2% (v/v) (3-Aminopropyl)triethoxysilane (APTES). React for 6 h at 70°C.
- Centrifuge, wash with ethanol, and dry under vacuum.
- Resuspend APTES-MSNs in MES buffer. Add PEI and EDC/NHS (molar ratio optimized for coating) and react for 4 h at room temperature.
- Wash thoroughly with water to obtain PEI-MSNs.
RNP Loading via Incubation:
- Incubate 1 mg of PEI-MSNs with 100 µL of 5 µM Cas9 RNP solution in nuclease-free buffer (e.g., PBS) for 12 hours at 4°C under gentle rotation.
- Centrifuge (10,000 rpm, 10 min) to collect RNP-loaded MSNs (MSN-RNP).
- Wash once gently to remove surface-adsorbed RNP. Determine loading efficiency by measuring RNP concentration in the supernatant (UV absorbance at 280 nm).
Transfection:
- Resuspend MSN-RNP complexes in serum-free medium.
- Add to cells (e.g., HEK293T) at a final concentration of 20 µg MSN/mL.
- Incubate for 4-6 hours, then replace with complete growth medium.
- Analyze editing efficiency 48-72 hours post-transfection.
Visualizations
Title: Workflow for CRISPR RNP Delivery Using Inorganic Nanoparticles
Title: Comparison of AuNP and Silica RNP Delivery Strategies
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Inorganic Nanoparticle-Mediated CRISPR-Cas9 RNP Delivery
| Reagent / Material | Supplier Examples | Function in Experimental Workflow |
|---|---|---|
| Chloroauric Acid (HAuCl₄) | Sigma-Aldrich, Thermo Fisher | Precursor for synthesis of gold nanoparticles (AuNPs). |
| Tetraethyl Orthosilicate (TEOS) | Sigma-Aldrich, Merck | Silicon alkoxide precursor for synthesizing silica nanoparticles. |
| (3-Aminopropyl)triethoxysilane (APTES) | Gelest, Sigma-Aldrich | Silane coupling agent for introducing amine groups on silica surfaces for further functionalization. |
| Methoxy-PEG-Thiol (MW 5000) | Creative PEGWorks, Laysan Bio | Provides stealth properties and stability to AuNPs; thiol group enables covalent gold-sulfur bonding. |
| Branched Polyethylenimine (PEI, 25 kDa) | Polysciences, Sigma-Aldrich | Cationic polymer used to coat nanoparticles, promoting endosomal escape via the "proton sponge" effect. |
| NHS-PEG-Maleimide | Thermo Fisher, Nanocs | Heterobifunctional crosslinker for covalent conjugation of targeting ligands (via thiols) to amine-functionalized nanoparticles. |
| Pure Cas9 Nuclease, S. pyogenes | IDT, Thermo Fisher, NEB | Core protein component for assembly of the Cas9 RNP complex in vitro. |
| Synthetic sgRNA (CRISPR RNA) | Synthego, Dharmacon, IDT | Guides the Cas9 protein to the specific genomic DNA target sequence. |
| CellTiter-Glo Luminescent Viability Assay | Promega | Quantifies cellular viability/cytotoxicity after nanoparticle treatment. |
| T7 Endonuclease I (T7E1) or ICE Analysis Tool | NEB, Synthego | Enzymatic assay (or computational analysis) for quantifying indel mutation frequency (editing efficiency). |
Within the context of CRISPR-Cas9 ribonucleoprotein (RNP) delivery via nanoparticles, surface functionalization with targeting ligands is a critical step to achieve cell-specific delivery, enhance cellular uptake, and reduce off-target effects. This Application Note details contemporary strategies for conjugating peptides, antibodies, and aptamers to nanoparticle surfaces, focusing on methodologies relevant to RNP-loaded nanocarriers.
Targeted Nanoparticle Design: Ligand Comparison
Table 1: Comparison of Targeting Ligands for RNP-Nanoparticle Conjugation
| Ligand Type | Size (kDa) | Binding Affinity (Kd) | Conjugation Chemistry | Key Advantages | Key Challenges for RNP Delivery |
|---|---|---|---|---|---|
| Peptides | 1-10 | µM - nM | NHS-Ester, Click Chemistry, Maleimide | Small size, high stability, low immunogenicity | Potential lower specificity, susceptibility to proteolysis |
| Antibodies | ~150 | nM - pM | NHS-Ester, Maleimide, Site-specific (e.g., Glycan) | Extremely high specificity and affinity | Large size can hinder penetration, immunogenicity, batch variability |
| Aptamers | 10-30 | nM - pM | Thiol-Maleimide, Click Chemistry, NHS-Ester | Small size, chemical stability, in vitro selection | Susceptible to nuclease degradation, potential renal clearance |
Detailed Application Protocols
Protocol 1: Conjugation of RGD Peptides to Lipid Nanoparticles (LNPs) via Maleimide-Thiol Chemistry
Application: Targeting integrin αvβ3-overexpressing cells (e.g., tumor endothelial cells) for Cas9 RNP delivery.
Materials (Research Reagent Solutions):
- LNP Formulation: Pre-formed, thiolated (SH-LNP) nanoparticles encapsulating Cas9 RNP.
- Targeting Ligand: cRGDfK peptide with C-terminal cysteine (e.g., Cyclo(Arg-Gly-Asp-D-Phe-Lys(Cys)).
- Conjugation Buffer: 0.1 M Phosphate, 1 mM EDTA, pH 6.5-7.2 (degassed, N₂ sparged).
- Traut's Reagent (2-iminothiolane): For introducing thiols if not present.
- PD-10 Desalting Columns: For purification.
Procedure:
- LNP Thiol Activation: If LNPs lack surface thiols, incubate with a 20-fold molar excess of Traut's Reagent in conjugation buffer for 1 hr at RT. Purify via size-exclusion chromatography (SEC) using PD-10 column equilibrated with degassed conjugation buffer.
- Ligand Preparation: Dissolve cRGDfK-Cys peptide in degassed conjugation buffer to 5 mM.
- Conjugation: Add a 1.5-2 molar excess of peptide solution to the SH-LNP suspension. React under gentle agitation for 4-6 hours at 4°C under inert atmosphere (N₂/Ar).
- Quenching & Purification: Add a 10-fold molar excess of N-ethylmaleimide (NEM) to quench unreacted thiols. Incubate 15 min. Purify conjugated LNPs (cRGD-LNPs) via SEC (e.g., Sepharose CL-4B) or tangential flow filtration using 1x PBS, pH 7.4.
- Validation: Determine conjugation efficiency using Ellman's assay for residual thiols and HPLC analysis of free peptide.
Protocol 2: Site-Specific Conjugation of Antibody Fragments (Fab') to Polymeric Nanoparticles via Click Chemistry
Application: Targeting cell-specific surface antigens with minimal steric hindrance for RNP delivery.
Materials (Research Reagent Solutions):
- Nanoparticles: Dibenzocyclooctyne (DBCO)-functionalized, RNP-loaded PLGA nanoparticles.
- Targeting Ligand: Fab' fragment with a terminal azide group (N₃-Fab').
- Reaction Buffer: 1x PBS, pH 7.4.
- Purification Device: 300 kDa MWCO centrifugal filters.
Procedure:
- Nanoparticle Preparation: Synthesize or purchase PLGA nanoparticles with surface DBCO groups (e.g., using DBCO-PEG-PLGA copolymer).
- Ligand Preparation: Ensure N₃-Fab' is in PBS, pH 7.4. Centrifuge briefly to remove aggregates.
- Click Conjugation: Mix N₃-Fab' solution with DBCO-NP suspension at a 50:1 molar ratio (Fab':NP). Incubate with gentle rotation for 24 hours at 4°C.
- Purification: Wash the reaction mixture 3x using 300 kDa MWCO centrifugal filters with PBS to remove unreacted Fab'.
- Validation: Analyze by SDS-PAGE (Coomasie stain) of nanoparticle-bound proteins and measure hydrodynamic diameter/zeta potential shift via Dynamic Light Scattering (DLS).
Protocol 3: Aptamer Functionalization of Gold Nanoparticles (AuNPs) for RNP Complexation via Thiol-Gold Binding
Application: Creating a stable, targeted complex for RNP delivery to specific cell types.
Materials (Research Reagent Solutions):
- AuNPs: 20 nm citrate-capped gold nanoparticles.
- Targeting Ligand: Thiol-modified DNA aptamer (e.g., anti-EGFR aptamer).
- Salting Buffer: 1x PBS containing 0.1% SDS, pH 7.4.
- TCEP Solution: Tris(2-carboxyethyl)phosphine, for reducing disulfide bonds.
- RNP Complex: Pre-formed Cas9 RNP with a cationic polymer coating (e.g., PEI).
Procedure:
- Aptamer Reduction: Incubate thiol-aptamer (100 µM) with 10 mM TCEP for 1 hr at RT. Purify via desalting column.
- Aptamer Immobilization: Add reduced aptamer (final conc. 2 µM) to AuNPs. Incubate for 1 hr. Gradually increase salt concentration to 0.1 M NaCl over 2 hours to stabilize AuNPs. Incubate overnight.
- Passivation: Add 10 µM mercapto-PEG to block remaining gold surface for 4 hrs.
- Purification: Centrifuge aptamer-AuNPs at 14,000 x g for 30 min. Resuspend in nuclease-free water.
- RNP Complexation: Mix aptamer-AuNPs with cationic polymer-coated RNP at a 1:10 weight ratio (AuNP:polymer). Incubate 30 min at RT to form the final delivery complex via electrostatic interaction.
- Validation: Use UV-Vis spectroscopy to confirm aptamer loading (shift in λmax). Validate targeting via cell-binding assays (e.g., flow cytometry).
Visualization
Diagram Title: Nanoparticle Targeting Ligand Conjugation & Uptake Path
Diagram Title: Surface Functionalization Protocol Workflow
The Scientist's Toolkit
Table 2: Essential Reagents for Ligand Conjugation in RNP Delivery
| Reagent / Material | Supplier Examples | Function in Conjugation |
|---|---|---|
| Maleimide-PEG-NHS Ester | Thermo Fisher, Sigma-Aldrich, Creative PEGWorks | Heterobifunctional crosslinker for amine-to-thiol coupling (e.g., antibody/peptide to NP). |
| DBCO-PEG-NHS Ester | Click Chemistry Tools, Sigma-Aldrich | Enables bioorthogonal copper-free click chemistry between DBCO and azide groups. |
| Traut's Reagent (2-Iminothiolane) | Thermo Fisher, TCI Chemicals | Introduces sulfhydryl (-SH) groups onto primary amines for thiol-based chemistry. |
| TCEP Hydrochloride | GoldBio, Thermo Fisher | Reduces disulfide bonds in antibodies/aptamers to generate free thiols for conjugation. |
| SM(PEG)₂₄ (Succinimidyl PEG) | Thermo Fisher | Creates a long, flexible PEG spacer arm to reduce steric hindrance of conjugated ligands. |
| HiTrap Desalting Columns | Cytiva | Rapid buffer exchange and removal of unreacted small molecules from nanoparticle suspensions. |
| Amicon Ultra Centrifugal Filters | MilliporeSigma | Concentrates and purifies nanoparticle-ligand conjugates based on size (MWCO selection). |
| Zetasizer Nano ZSP | Malvern Panalytical | Characterizes hydrodynamic size, polydispersity (PDI), and zeta potential of functionalized NPs. |
| MicroBCA Protein Assay Kit | Thermo Fisher | Quantifies the amount of protein (antibody/peptide) conjugated to the nanoparticle surface. |
In the development of lipid nanoparticles (LNPs) for CRISPR-Cas9 ribonucleoprotein (RNP) delivery, comprehensive physicochemical and biological characterization is paramount. This application note details standardized protocols for four critical assays: particle size and polydispersity (PDI), zeta potential, encapsulation efficiency (EE%), and RNP integrity. These parameters directly dictate the stability, cellular uptake, endosomal escape, and ultimately, the gene editing efficacy of the LNP-RNP formulation. The protocols are framed within a thesis investigating the structure-activity relationship of ionizable lipids in RNP delivery.
Research Reagent Solutions & Essential Materials
| Item | Function/Brief Explanation |
|---|---|
| Malvern Panalytical Zetasizer Ultra | Dynamic Light Scattering (DLS) for size/PDI; Electrophoretic Light Scattering for zeta potential. |
| 1x PBS, pH 7.4 (0.1x dilution) | Standard low-ionic-strength aqueous dispersant for DLS and zeta potential measurements. |
| Disposable Folded Capillary Cells (DTS1070) | Cuvettes for zeta potential measurement, suitable for aqueous electrophoretic mobility. |
| SYBR Gold Nucleic Acid Gel Stain | Fluorescent dye for quantifying unencapsulated, accessible guide RNA (gRNA). |
| Quant-iT RiboGreen RNA Assay Kit | Ultra-sensitive fluorescent assay for total (free + encapsulated) RNA quantification. |
| Triton X-100 (1% v/v) | Non-ionic detergent used to lyse LNPs and release encapsulated cargo for total RNA measurement. |
| Native PAGE Gel (4-20% Tris-Glycine) | For assessing Cas9-gRNA RNP complex formation and integrity post-encapsulation/release. |
| Recombinant S. pyogenes Cas9 Nuclease | Core enzyme component for forming the RNP complex with in vitro-transcribed or synthetic gRNA. |
| Ionizable Lipid (e.g., DLin-MC3-DMA, SM-102) | Key LNP component that protonates in acidic endosomes, enabling membrane disruption and RNP release. |
| MicroBCA Protein Assay Kit | For quantifying total Cas9 protein, used in conjunction with RNA assays to calculate RNP-specific EE. |
Protocols & Data Presentation
Particle Size and Polydispersity Index (PDI) by Dynamic Light Scattering (DLS)
Protocol:
- Dilute the purified LNP-RNP formulation 1:50 in 1x PBS, pH 7.4, to achieve an optimal scattering intensity.
- Equilibrate the sample and dispersant (PBS) to 25°C for 120 seconds in the Zetasizer sample chamber.
- Load 1 mL of diluted sample into a disposable polystyrene cuvette.
- Perform measurement using the following parameters:
- Measurement Angle: Backscatter (173°)
- Number of Runs: 3 measurements of 13-15 sub-runs each
- Viscosity & Refractive Index: Use pre-set values for water.
- Analyze results using the "Multiple Narrow Modes" or "General Purpose" analysis model within the Zetasizer Software.
Table 1: Representative LNP-RNP Size and PDI Data
| Formulation ID | Ionizable Lipid | Z-Average (d.nm) | PDI | Interpretation |
|---|---|---|---|---|
| LNP-RNP-A | DLin-MC3-DMA | 84.2 ± 3.1 | 0.08 ± 0.02 | Monodisperse, optimal for delivery. |
| LNP-RNP-B | SM-102 | 92.7 ± 5.4 | 0.11 ± 0.03 | Narrow distribution, suitable. |
| LNP-RNP-C | Novel Lipid X | 152.3 ± 12.7 | 0.23 ± 0.05 | Polydisperse; may indicate instability. |
Zeta Potential by Electrophoretic Light Scattering
Protocol:
- Dilute LNP-RNP formulation 1:100 in 0.1x PBS (low conductivity) or 1 mM NaCl.
- Rinse a folded capillary cell 2-3 times with the diluted sample, then fill it completely.
- Insert the cell into the Zetasizer and equilibrate to 25°C.
- Set measurement parameters:
- Smoluchowski Model: Applied (for aqueous dispersions).
- Number of Measurements: Minimum 3, with >12 sub-runs each.
- Voltage Selection: Automatic.
- The software calculates zeta potential from the measured electrophoretic mobility.
Table 2: Representative Zeta Potential Data
| Formulation ID | Dispersion Medium | Zeta Potential (mV) | Interpretation |
|---|---|---|---|
| LNP-RNP-A | 1 mM NaCl | -2.1 ± 0.8 | Near-neutral surface charge, typical for PEGylated LNPs. |
| LNP-RNP-B | 1 mM NaCl | -3.5 ± 1.2 | Slight negative charge, suggests minimal aggregation risk. |
| "Empty" LNPs | 1 mM NaCl | +5.2 ± 1.5 | Positive charge from ionizable lipid headgroups when empty. |
Encapsulation Efficiency (EE%) by Fluorometric RNA Assay
Protocol (Dual-Method for RNP): This protocol quantifies both RNA and protein encapsulation to confirm RNP co-encapsulation.
- A. gRNA Encapsulation (RiboGreen Assay):
- Prepare two sets of samples in a black 96-well plate in triplicate: "Total" (LNPs lysed with 1% Triton X-100) and "Free" (LNPs in PBS only).
- Dilute samples 1:200 in TE buffer (200 µL final volume).
- Add 100 µL of Quant-iT RiboGreen reagent (1:1000 dilution in TE) to each well.
- Incubate protected from light for 5 minutes.
- Measure fluorescence (ex/em ~480/520 nm). Calculate gRNA concentration from an RNA standard curve (0-100 ng/mL).
- B. Cas9 Protein Encapsulation (MicroBCA Assay):
- Similarly, prepare "Total" (Triton-lysed) and "Free" (intact LNPs) sample sets.
- Follow the manufacturer's protocol for the MicroBCA assay.
- Measure absorbance at 562 nm. Calculate Cas9 concentration from a BSA standard curve.
- Calculation:
EE% (for RNA or Protein) = [1 - (Concentration in "Free" Sample / Concentration in "Total" Sample)] x 100
Table 3: Representative Encapsulation Efficiency Data
| Formulation ID | gRNA EE% (RiboGreen) | Cas9 EE% (MicroBCA) | RNP Co-Encapsulation Ratio (gRNA/Cas9) |
|---|---|---|---|
| LNP-RNP-A | 95.4% ± 2.1% | 93.8% ± 3.4% | 1.02:1 |
| LNP-RNP-B | 88.7% ± 4.5% | 85.2% ± 5.1% | 1.04:1 |
| LNP (Free gRNA) | 70.3% ± 8.2% | N/A | N/A |
RNP Integrity Assay by Native PAGE
Protocol:
- Release RNP: Treat LNP-RNP formulation with 1% Triton X-100 and incubate at room temperature for 15 min.
- Prepare Samples: Mix released cargo 1:1 with 2x Native Sample Buffer (no SDS, no reducing agents). Include controls: Naked RNP complex, free Cas9, free gRNA.
- Electrophoresis: Load samples onto a pre-chilled 4-20% Tris-Glycine native gel. Run in 1x Tris-Glycine Native Running Buffer at 100V for 90-120 minutes at 4°C.
- Staining: Stain for protein using Coomassie Blue to visualize Cas9. Subsequently, stain for nucleic acid using SYBR Gold to visualize gRNA.
- Analysis: Gel imaging should show co-migration of the Cas9 band (Coomassie) and the gRNA band (SYBR Gold) at the same molecular weight as the naked RNP control, indicating intact complex.
Experimental Workflow & Data Interpretation Diagrams
Title: LNP-RNP Critical Characterization Workflow
Title: Physicochemical Traits Drive Editing Efficiency
Within the broader thesis on CRISPR-Cas9 ribonucleoprotein (RNP) delivery via engineered nanoparticles, the development of robust, cell line-optimized in vitro transfection protocols is a foundational step. The efficacy of RNP-mediated genome editing is inherently constrained by the efficiency of intracellular delivery, which varies dramatically across cell types due to differences in physiology, division rate, and endocytic machinery. This document provides detailed application notes and protocols for cell line-specific RNP transfection, emphasizing efficiency metrics critical for downstream analysis in therapeutic development.
Cell Line-Specific Considerations
The success of nanoparticle-mediated RNP delivery is highly dependent on the target cell line. Key biological variables must be assessed prior to protocol design.
- Proliferation Rate: Actively dividing cells often show higher CRISPR editing efficiency, partly due to increased nuclear membrane permeability during mitosis. Slow-growing or primary cells present a significant challenge.
- Endocytic Pathways: Different cell lines preferentially utilize distinct endocytic pathways (e.g., clathrin-mediated, caveolin-mediated, macropinocytosis). Nanoparticle surface chemistry must be tailored to exploit the dominant pathway for efficient endosomal escape.
- Cell Surface Proteoglycans: The abundance of heparan sulfate proteoglycans (HSPGs) can influence the binding and uptake of cationic or charged nanoparticles.
- Innate Immune Response: Some cell lines, particularly primary and stem cells, may exhibit pronounced inflammatory responses to foreign nucleic acids or carrier materials, affecting viability and editing outcomes.
The following table summarizes critical parameters for common cell lines used in CRISPR-Cas9 RNP research.
Table 1: Cell Line Characteristics Relevant to Nanoparticle RNP Transfection
| Cell Line | Cell Type | Typical Doubling Time | Recommended Seeding Density (per well in 24-plate) | Relative Transfection Difficulty | Dominant Endocytic Pathway | Notes for RNP Delivery |
|---|---|---|---|---|---|---|
| HEK293T | Human Embryonic Kidney | ~24 hours | 5.0 x 10⁴ | Easy | Clathrin-mediated | High division rate and high transfection efficiency. A standard model for protocol optimization. |
| HeLa | Human Cervical Carcinoma | ~24 hours | 4.5 x 10⁴ | Easy | Clathrin-mediated | Robust, readily transfected. Good for proof-of-concept RNP delivery studies. |
| HepG2 | Human Hepatocellular Carcinoma | ~48 hours | 6.0 x 10⁴ | Moderate | Multiple (Clathrin & Caveolin) | Slower growth. Can be sensitive to lipid-based transfection reagents. |
| U2OS | Human Osteosarcoma | ~30 hours | 4.0 x 10⁴ | Moderate | Caveolin-mediated | Useful for studying DNA repair pathways post-RNP cleavage. |
| hIPS Cells | Human Induced Pluripotent Stem | ~36 hours | 1.0 x 10⁵ | High / Difficult | Macropinocytosis | Sensitive to cytotoxicity. Requires gentle, optimized reagents. Low uptake efficiency common. |
| Primary T Cells | Human Primary Immune Cells | Variable (activated) | 5.0 x 10⁵ | High / Difficult | Variable | Highly sensitive to reagent toxicity. Often requires electroporation, but nanoparticle delivery is an active area of research. |
Detailed Protocol: Lipid-Based Nanoparticle (LNP) Transfection of CRISPR-Cas9 RNP in HEK293T and HeLa Cells
Materials & Reagent Solutions
Table 2: Research Reagent Solutions Toolkit
| Item | Function/Description |
|---|---|
| Cas9 Nuclease, purified | The effector protein for DNA cleavage. Must be high-purity, nuclease-free. |
| sgRNA (chemically synthesized) | Guides the Cas9 protein to the specific genomic locus. Reconstituted in nuclease-free duplex buffer. |
| Commercial LNP Transfection Reagent (e.g., Lipofectamine CRISPRMAX) | Lipid-based nanoparticles formulated specifically for RNP delivery. Provides encapsulation and promotes endosomal escape. |
| Opti-MEM I Reduced Serum Medium | Low-serum medium used for diluting nanoparticles and RNP complexes to prevent interference with formation. |
| Complete Growth Medium | Cell line-specific medium (e.g., DMEM + 10% FBS) for cell maintenance and post-transfection recovery. |
| Genomic DNA Extraction Kit | For harvesting genomic DNA post-transfection to assess editing efficiency. |
| T7 Endonuclease I (or similar) | Surveyor nuclease for detecting insertion/deletion (indel) mutations via mismatch cleavage assay. |
| Flow Cytometry Buffer (PBS + 2% FBS) | For processing cells for viability or co-transfected reporter assays. |
Method
Day 1: Cell Seeding
- Harvest HEK293T or HeLa cells in mid-log phase.
- Count cells and dilute in complete growth medium without antibiotics.
- Seed a 24-well plate at a density of 5.0 x 10⁴ cells per well in 500 µL of medium. Swirl gently to ensure even distribution.
- Incubate cells overnight at 37°C, 5% CO₂ to achieve ~70-80% confluency at the time of transfection.
Day 2: RNP Complex Formation & Transfection
- Prepare Cas9 RNP Complex:
- In a sterile microcentrifuge tube, combine 2.5 µg of purified Cas9 protein with 1.0 µg of target-specific sgRNA (molar ratio ~1:2).
- Add nuclease-free water to a total volume of 25 µL.
- Mix gently by pipetting and incubate at room temperature for 10-20 minutes to allow RNP complex formation.
- Prepare Lipid Nanoparticle (LNP) Mixture:
- In a separate tube, dilute 2.0 µL of LNP transfection reagent in 25 µL of Opti-MEM. Mix gently.
- Incubate at room temperature for 5 minutes.
- Form Final Transfection Complexes:
- Combine the 25 µL of prepared RNP complex with the 25 µL of diluted LNP reagent (total volume = 50 µL).
- Mix by gentle pipetting. Do not vortex.
- Incubate the combined mixture at room temperature for 10-15 minutes to allow nanoparticle encapsulation of the RNP.
- Transfect Cells:
- Add the 50 µL transfection complex dropwise to the wells containing seeded cells and 500 µL medium. Gently rock the plate.
- Return the plate to the 37°C, 5% CO₂ incubator.
Day 3: Medium Change (Optional)
- 4-6 hours post-transfection, carefully aspirate the transfection medium and replace with 500 µL of fresh, pre-warmed complete growth medium. This step can improve viability for sensitive cell lines.
Day 4-5: Harvest and Analysis
- Harvest cells 72 hours post-transfection for genomic DNA extraction and analysis of editing efficiency.
Efficiency Metrics and Analysis Protocols
A multi-parametric approach is required to fully evaluate transfection success.
1. Cell Viability Assessment (MTT or Flow Cytometry)
- Protocol: 72 hours post-transfection, add MTT reagent (0.5 mg/mL final concentration) to cells. Incubate 3-4 hours. Solubilize formazan crystals with DMSO and measure absorbance at 570 nm. Normalize to untreated control cells.
- Metric: % Viability = (Abssample / Abscontrol) * 100.
2. Transfection/Delivery Efficiency (Using Fluorescently Labeled RNP)
- Protocol: Use Cas9 protein conjugated to a fluorophore (e.g., Alexa Fluor 555) during the RNP formation step. 24 hours post-transfection, analyze cells by flow cytometry.
- Metric: % Delivery Efficiency = (Fluorescent-positive cell count / Total cell count) * 100.
3. Genome Editing Efficiency (T7 Endonuclease I Assay)
- Protocol:
- Extract genomic DNA from harvested cells.
- PCR-amplify the target genomic region (amplicon size: 400-800 bp).
- Hybridize and re-anneal PCR products: Denature at 95°C for 10 min, then ramp down to 25°C at -0.3°C/sec.
- Digest re-annealed products with T7E1 enzyme (NEB) for 30-60 min at 37°C.
- Run digested products on a 2% agarose gel.
- Metric Calculation:
- Quantify band intensities using gel analysis software (e.g., ImageJ).
- Apply the formula: % Indel = 100 * (1 - sqrt(1 - (b + c)/(a + b + c))), where a is the intensity of the undigested PCR product, and b & c are the intensities of the cleavage products.
Table 3: Typical Efficiency Metrics by Cell Line for LNP-RNP Delivery
| Cell Line | Expected Viability (% of Ctrl) | Expected Delivery Efficiency (%) | Expected Indel Efficiency (%) | Key Optimization Parameter |
|---|---|---|---|---|
| HEK293T | 85-95% | 80-95% | 60-80% | RNP:LNP ratio; complexation time. |
| HeLa | 80-90% | 75-90% | 50-75% | Cell confluence at transfection. |
| HepG2 | 70-85% | 60-80% | 30-60% | Use of serum-free during complexation; post-transfection medium change timing. |
| hIPS Cells | 60-75% | 40-70% | 20-50% | Seeding density; use of ROCK inhibitor; specialized LNPs. |
Visualizations
RNP-LNP Transfection Workflow
Intracellular RNP Delivery Pathway
Overcoming Hurdles: Strategies to Maximize Efficiency, Specificity, and Safety
Abstract: A primary bottleneck in CRISPR-Cas9 ribonucleoprotein (RNP) delivery via nanoparticles is low observed gene editing efficiency. This Application Note provides a structured diagnostic framework to determine if inefficiency stems from inadequate cellular internalization or from failure of cytosolic release of the RNP. We present quantitative assays, comparative data tables, and step-by-step protocols to enable researchers to isolate and address the limiting step.
Diagnostic Framework: Key Questions & Assays
| Hypothesized Limitation | Diagnostic Question | Primary Quantitative Assays |
|---|---|---|
| Poor Cellular Uptake | Is the nanoparticle-RNP complex efficiently internalized by target cells? | Flow Cytometry (Fluorophore-labeled RNP/Cas9) Confocal Microscopy (Co-localization with endosomal markers) |
| Deficient Cytosolic Release | Are internalized RNPs successfully escaping endo/lysosomal compartments? | Fluorescence Co-localization & Quenching Assay (e.g., Dye/Quencher RNP) Gal8/GFP Recruitment Assay (for endosomal damage) Functional Editing vs. Internalization Correlation |
Table 1: Expected Data Ranges for Key Diagnostic Assays
| Assay | Metric | Indicative of Poor Uptake | Indicative of Successful Uptake but Poor Release |
|---|---|---|---|
| Flow Cytometry | % Fluorescence-positive cells | < 70% (for high MOI delivery) | > 85% |
| Median Fluorescence Intensity (MFI) | Low MFI relative to dose | High MFI | |
| Confocal Co-localization (Pearson's Coefficient) | RNP vs. Early Endosome (EEA1) | Low coefficient (<0.4) | High coefficient (>0.7) at early timepoints, persistent over time |
| RNP vs. Lysosome (LAMP1) | Low coefficient | Very high coefficient (>0.8) at late timepoints (24h) | |
| Gal8/GFP Recruitment Assay | % of Gal8 puncta-positive cells | Not applicable | < 20% (suggests minimal endosomal disruption) |
| Fluorophore-Quencher Assay | Cytosolic fluorescence signal ratio | Not applicable | Low signal ratio (< 2-fold increase post-lysis) |
Experimental Protocols
Protocol 1: Quantifying Cellular Uptake via Flow Cytometry
Objective: Distinguish surface-bound from internalized nanoparticle-RNP complexes. Key Reagents: Fluorescently labeled Cas9 protein (e.g., Alexa Fluor 647-Cas9), appropriate nanoparticle formulation, target cells.
- Complex Formation: Formulate nanoparticles with labeled Cas9 RNP per standard protocol.
- Dose & Incubate: Treat cells with formulated RNPs (e.g., 50-200nM RNP equivalent) for 4-6h at 37°C.
- Surface Stripping: To remove surface-bound particles, wash cells with cold PBS, then treat with 0.25% Trypsin-EDTA for 5 min at 37°C, or use an acid wash (0.1M Glycine, pH 3.0) for 1 min. Neutralize immediately.
- Analysis: Harvest cells, wash with PBS, and analyze via flow cytometry. Gate on live cells. Critical Control: Include samples incubated at 4°C (inhibits endocytosis) to confirm signal is from internalization.
Protocol 2: Assessing Cytosolic Release via Dye/Quencher RNP Assay
Objective: Detect the translocation of RNPs from quenched endosomes into the cytosol. Key Reagents: Cas9 RNP labeled with a pH-insensitive fluorophore (e.g., Alexa Fluor 647) and conjugated to a quencher (e.g., QSY21) via a cleavable linker (e.g., disulfide bond), reducing agent (e.g., DTT).
- Probe Preparation: Conjugate quencher to labeled RNP using a disulfide linker. Purify the quenched RNP.
- Cell Treatment: Treat cells with nanoparticle/quenched-RNP complexes as usual.
- Live-Cell Imaging/Flow Analysis:
- Timepoints: Image or analyze cells at 2, 4, 8, 24h post-treatment.
- Principle: While in endosomes, fluorescence remains quenched. Upon cytosolic release, the reducing environment cleaves the disulfide bond, separating fluorophore and quencher, restoring fluorescence.
- Quantification: Measure fluorescence intensity in live cells. Key Validation: Lyse cells at endpoint with 10mM DTT to measure total recoverable fluorescence. The cytosolic release efficiency is proportional to the ratio of live-cell signal to post-lysis signal.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent/Material | Function in Diagnosis | Example Product/Note |
|---|---|---|
| Fluorescently Labeled Cas9 | Enables tracking of RNP uptake and localization via microscopy/flow cytometry. | Label with amine-reactive dyes (e.g., Alexa Fluor 488/647). Ensure labeling does not impair RNP activity. |
| Endosomal/Lysosomal Markers | Antibodies or fluorescent conjugates to mark specific compartments for co-localization studies. | Anti-EEA1 (Early Endosomes), Anti-LAMP1 (Lysosomes), LysoTracker dyes. |
| Galectin-8 (Gal8) Reporter | A cell line expressing Gal8 fused to GFP or mCherry. Gal8 binds exposed glycans upon endosomal damage, forming visible puncta. | U2OS Gal8-mGFP reporter cell line. A robust indicator of endosomal disruption/leakage. |
| Fluorophore-Quencher Pair | For constructing the cytosolic release sensor RNP. | e.g., Alexa Fluor 647 (fluorophore) & QSY21 (quencher). Link via a reducible SS-linker. |
| Polymer/Lipid Nanoparticles | Standardized delivery vehicle controls for benchmarking. | Commercial lipid nanoparticles (e.g., Lipofectamine CRISPRMAX) or well-characterized polymeric NPs (e.g., polyplexes). |
| Chemical Release Enhancers | Control reagents to artificially induce endosomal release for validation. | Chloroquine (lysosomotropic agent), Bafilomycin A1 (V-ATPase inhibitor). Use as positive controls for release assays. |
Diagnostic Decision Pathways & Workflows
Diagram 1: Diagnostic Decision Tree for Low Editing Efficiency
Diagram 2: Mechanism of the Dye-Quencher Cytosolic Release Assay
A principal challenge in therapeutic CRISPR-Cas9 RNP delivery via nanoparticles is the endosomal entrapment and subsequent lysosomal degradation of the cargo. Efficient endosomal escape is a critical rate-limiting step. This document details contemporary chemical and material strategies designed to overcome this bottleneck, framed within ongoing research on lipid and polymeric nanoparticle (LNP, PNP) platforms for RNP delivery.
Application Notes: Mechanisms & Quantitative Comparisons
Key Mechanisms of Action
- Proton-Sponge Effect: Polymers with high buffering capacity in the endosomal pH range (7.4-5.0) absorb incoming protons, causing an influx of chloride ions and water. The resulting osmotic swelling ruptures the endosome.
- Membrane Fusion/Destabilization: Fusogenic lipids or peptides undergo conformational changes at low pH, inserting into and destabilizing the endosomal lipid bilayer to create pores or induce fusion.
- Pore Formation: Certain materials, like some synthetic peptides, can assemble into transmembrane pores.
Table 1: Comparison of Endosomal Escape Agents in CRISPR-Cas9 RNP Delivery Systems
| Escape Agent Class | Example Materials | Typical Loading (mol%)/Concentration | Reported Escape Efficiency | Key Advantages | Noted Cytotoxicity |
|---|---|---|---|---|---|
| Proton-Sponge Polymers | Branched PEI (25 kDa), PBAE, PLL-g-DEX | 50-70 wt% of polymer core | 15-35% (per endosome) | High buffering, design versatility | Medium-High (depends on Mw, structure) |
| Fusogenic Lipids | DOPE, DOSPA, CLEM-1 | 20-50 mol% of lipid bilayer | 20-40% (per endosome) | Biocompatible, biomimetic | Low-Medium |
| pH-Sensitive Peptides | GALA, INF7, HA2 derivatives | 5-15 mol% of formulation | 10-30% (per endosome) | High specificity, low immunogenicity | Low |
| Porphyrin-Based | TPP, Zn-Por | 1-5 mol% of lipid bilayer | 25-50% (via photochemical) | Spatiotemporally controllable | Medium (photo-dependent) |
Table 2: Performance Metrics of RNP Nanoparticles Incorporating Escape Agents
| Nanoparticle Formulation | Escape Agent | Cell Model | Gene Editing Efficiency (%) | Endosomal Escape Half-time (min) | Citation (Example) |
|---|---|---|---|---|---|
| Cationic Lipid Nanoassembly | DOPE/CLEM-1 | HeLa | ~45 | ~15-20 | Liu et al., 2023 |
| PEG-PBAE Hybrid NP | PBAE (pH-sensitive) | Primary T-cells | ~60 | ~10-15 | Liu et al., 2023 |
| Charge-Averting LNP | DOPE/DLin-MC3-DMA | Hepatocytes (in vivo) | ~30 (in vivo) | N/A | Wei et al., 2021 |
| Gold-Nucleic Acid Nanocomplex | HA2 peptide conjugate | U2OS | ~55 | ~20 | Ruan et al., 2022 |
Experimental Protocols
Protocol: Evaluating Endosomal Escape Using a Split GFP Complementation Assay
Objective: Quantitatively measure endosomal escape kinetics and efficiency of CRISPR-Cas9 RNP-loaded nanoparticles. Principle: Cas9 is fused to GFP11 tag. Cells express GFP1-10. Functional GFP fluorescence only complements upon cytosolic delivery of Cas9-GFP11.
Materials:
- Stable cell line expressing GFP1-10.
- Purified Cas9-GFP11 fusion protein + sgRNA (RNP).
- Test nanoparticles (e.g., DOPE-containing LNPs, PEI-based PNPs).
- Control nanoparticles (no escape agent).
- Live-cell imaging setup with environmental control.
Procedure:
- Seed Cells: Plate GFP1-10 cells in glass-bottom dishes 24h pre-transfection.
- Prepare RNP-NPs: Formulate nanoparticles encapsulating Cas9-GFP11 RNP per standard protocols (e.g., microfluidics for LNPs, co-complexation for polyplexes). Include escape agent in test formulations.
- Treat Cells: Replace medium with pre-warmed imaging medium. Add nanoparticle dose (e.g., 50 nM RNP equivalent).
- Live-Cell Imaging: Immediately place dish on confocal microscope (37°C, 5% CO₂). Acquire images every 5 minutes for 2-4 hours at 488 nm excitation.
- Data Analysis:
- Kinetics: Plot mean cytosolic fluorescence intensity over time for ≥50 cells per condition.
- Efficiency: Calculate the percentage of cells exhibiting cytosolic fluorescence above a defined threshold at the 2-hour time point.
- Half-time: Determine time point at which 50% of maximal fluorescence is achieved.
Protocol: Formulating pH-Sensitive Polymeric Nanoparticles (PNPs) for RNP Delivery
Objective: Prepare and characterize branched PEI-based nanoparticles for Cas9 RNP delivery. Materials: Branched PEI (25 kDa), Cas9 protein, sgRNA, Sodium Acetate Buffer (25 mM, pH 5.0), Nuclease-free water, Hepes Buffered Saline (HBS, pH 7.4).
Procedure:
- RNP Complexation: Incubate Cas9 (final 2 µM) with sgRNA (1:1.2 molar ratio) in sodium acetate buffer for 10 min at RT.
- Polymer Solution: Dilute PEI in sodium acetate buffer to 1 mg/mL.
- Nanoparticle Formation: Rapidly mix equal volumes (e.g., 50 µL) of RNP solution and PEI solution via vortexing for 30 sec. Incubate for 30 min at RT for complex coacervation.
- Buffer Exchange: Dilute the mixture 1:5 in HBS (pH 7.4). Optionally, concentrate using a 100 kDa MWCO centrifugal filter.
- Characterization:
- Size/PDI/Zeta Potential: Measure via dynamic light scattering in HBS.
- RNP Encapsulation: Quantify using fluorescently labeled sgRNA and a Ribogreen assay post-purification.
- Morphology: Assess by transmission electron microscopy (negative stain).
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Studying Endosomal Escape in RNP Delivery
| Reagent/Material | Supplier Examples | Function/Utility |
|---|---|---|
| 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) | Avanti Polar Lipids, Sigma-Aldrich | Fusogenic lipid for pH-sensitive liposomal formulations. |
| Branched Polyethylenimine (PEI), 25 kDa | Polysciences, Sigma-Aldrich | Benchmark proton-sponge polymer for polyplex formation. |
| Endocytosis Inhibitors (Chlorpromazine, Dynasore) | Tocris, Sigma-Aldrich | Pharmacological tools to dissect uptake pathways pre-escape. |
| Lysotracker Red DND-99 | Thermo Fisher Scientific | Fluorescent dye to label acidic organelles (lysosomes/endosomes). |
| Split GFP System (GFP1-10 + GFP11) | Addgene, in-house generation | Gold-standard quantitative assay for cytosolic delivery. |
| pH-Sensitive Dye (e.g., pHrodo Dextran) | Thermo Fisher Scientific | Conjugate to nanoparticles to visualize endosomal acidification/rupture. |
| Poly(beta-amino ester) (PBAE) Library | Specific polymers from Sigma or synthesized | Customizable, biodegradable proton-sponge polymers. |
| GALA Peptide | Genscript, AnaSpec | Model pH-sensitive fusogenic peptide for conjugation. |
| Microfluidic Mixer (NanoAssemblr, iLiNP) | Precision NanoSystems | For reproducible, scalable production of LNPs/PNPs. |
Visualizations
Diagram Title: Proton-Sponge Mechanism for Endosomal Escape
Diagram Title: Workflow: Evaluating RNP Escape with Split-GFP
Within CRISPR-Cas9 RNP delivery research utilizing lipid nanoparticles (LNPs) or polymeric nanoparticles, the stability of the RNP cargo prior to encapsulation is a critical bottleneck. The efficacy of the final therapeutic nanoparticle hinges on the initial activity of the RNP complex. This application note details current strategies for formulating and stabilizing RNPs in solution for long-term storage, ensuring high activity for subsequent nanoparticle formulation and gene editing applications.
Key Stabilization Challenges and Strategies
RNPs are susceptible to aggregation, Cas9 protein denaturation, and guide RNA (gRNA) degradation via hydrolysis or RNase activity. Effective stabilizers address these issues through cryoprotection, steric hindrance, and thermodynamic stabilization.
Quantitative Data on Common Stabilizers and Buffers
Table 1: Efficacy of Common Additives in RNP Storage Buffers
| Additive Category | Specific Agent | Typical Concentration | Primary Function | Reported Residual Activity (vs. fresh)* |
|---|---|---|---|---|
| Polyols/Sugars | Trehalose | 5-10% (w/v) | Cryoprotectant, water replacement | >90% (6 months, -80°C) |
| Glycerol | 5-20% (v/v) | Cryoprotectant, reduces ice formation | ~85% (12 months, -80°C) | |
| Polymers | PEG-8000 | 0.01-0.1% (w/v) | Steric stabilization, prevents aggregation | ~88% (3 months, -20°C) |
| Surfactants | Poloxamer 188 | 0.001-0.01% (w/v) | Reduces surface-induced denaturation/adsorption | >90% (1 month, 4°C) |
| Reducing Agents | DTT | 1-5 mM | Maintains Cas9 cysteines in reduced state | Varies; can be detrimental long-term |
| RNase Inhibitors | SUPERase•In | 0.5-1 U/µL | Protects gRNA from degradation | >95% (1 month, -80°C) |
| Carrier Proteins | BSA or HSA | 0.1-1 mg/mL | Competes for surface adsorption, stabilizer | ~80% (6 months, -80°C) |
*Activity is measured by in vitro cleavage assays or cellular editing efficiency post-storage. Results are model-dependent.
Table 2: Comparison of Formulation Buffer Compositions for Long-Term Storage
| Buffer Component | Standard HEPES-Salt Buffer | Enhanced Stabilization Buffer | Lyophilization Buffer |
|---|---|---|---|
| Base Buffer | 20 mM HEPES, pH 7.5 | 20 mM HEPES, pH 7.5 | 20 mM Tris, pH 8.0 |
| Salt | 150 mM KCl | 150 mM KCl | 50 mM KCl |
| Polyol | - | 5% (w/v) Trehalose | 10% (w/v) Trehalose |
| Polymer/Surfactant | - | 0.01% Poloxamer 188 | 0.05% PEG-8000 |
| Carrier Protein | - | 0.1 mg/mL HSA | - |
| RNase Inhibitor | Optional | 0.5 U/µL | - |
| Recommended Storage | -80°C, short-term (weeks) | -80°C for >1 year | Lyophilized, -20°C |
| Key Advantage | Simple, low interference | Maximizes liquid-state stability | Enables ambient temp storage |
Detailed Experimental Protocols
Protocol 1: Formulating and Storing RNPs in Enhanced Stabilization Buffer Objective: To prepare a stable RNP complex suitable for long-term storage at -80°C prior to nanoparticle formulation. Materials: Purified Cas9 protein, synthetic gRNA, HEPES-KOH pH 7.5, KCl, Trehalose, Poloxamer 188, Human Serum Albumin (HSA), RNase inhibitor, filter tubes. Procedure:
- Prepare 5X Enhanced Stabilization Buffer (ESB): 100 mM HEPES pH 7.5, 750 mM KCl, 25% (w/v) trehalose, 0.05% (w/v) Poloxamer 188, 0.5 mg/mL HSA. Filter sterilize (0.22 µm).
- Complex Formation: Dilute Cas9 protein and gRNA to 2x final concentration in 1X dilution buffer (20 mM HEPES, 150 mM KCl). Mix equimolar amounts and incubate at 25°C for 10 min.
- Final Formulation: Add 1 volume of 5X ESB and 0.5 U/µL RNase inhibitor to 4 volumes of the formed RNP complex. Mix gently by pipetting.
- Aliquoting and Storage: Dispense into low-protein-binding tubes as single-use aliquots. Flash-freeze in liquid nitrogen and store at -80°C.
- Quality Control: Assess post-thaw activity via an in vitro DNA cleavage assay compared to a freshly prepared control.
Protocol 2: Assessing RNP Stability via In Vitro Cleavage Assay Objective: To quantitatively measure the residual DNA cleavage activity of stored RNP samples. Materials: Stored RNP aliquots, target plasmid DNA (containing target sequence), NEBuffer 3.1, Agarose, TAE buffer, gel imaging system. Procedure:
- Thaw RNP on ice and dilute to 100 nM in 1X ESB.
- Set Up Reaction: In a 20 µL volume, combine 200 ng of target plasmid, 1X NEBuffer 3.1, and a dilution series of RNP (e.g., 10, 25, 50 nM). Incubate at 37°C for 1 hour.
- Stop Reaction: Add Proteinase K and SDS to final 0.5 mg/mL and 0.1% respectively. Incubate at 55°C for 15 min.
- Analysis: Run samples on a 1% agarose gel. Quantify the bands corresponding to uncut (supercoiled/nicked) and cut (linear) plasmid using gel analysis software.
- Calculate Activity: Determine the concentration of RNP required for 50% cleavage (EC50). Compare the EC50 of the stored sample to that of a freshly prepared RNP standard. Report residual activity as (EC50fresh / EC50stored) * 100%.
The Scientist's Toolkit: Research Reagent Solutions
| Reagent/Material | Function & Importance |
|---|---|
| Trehalose (Diabetic) | Non-reducing disaccharide; forms stable glassy state, protects proteins via water replacement theory. Critical for lyophilization. |
| Poloxamer 188 (Pharma Grade) | Non-ionic triblock copolymer surfactant; reduces interfacial stress during freeze-thaw and prevents RNP adhesion to surfaces. |
| Recombinant Human Serum Albumin (rHSA) | Carrier protein; minimizes surface adsorption, provides colloidal stability, and is regulatory-friendly for therapeutic development. |
| SUPERase•In RNase Inhibitor | Broad-spectrum RNase inhibitor; actively degrades RNases, superior to RNasin for protecting single-guide RNA in long-term storage. |
| Low-Binding Microcentrifuge Tubes | Surface-treated polypropylene; minimizes loss of low-concentration RNP complexes via adsorption. |
| HEPES Buffer (Ultra Pure) | Biological buffer with minimal metal ion chelation; maintains stable pH during freeze-thaw cycles compared to Tris. |
| Size-Exclusion Spin Columns (e.g., Zeba) | Allows rapid buffer exchange into final stabilization buffer, removing contaminants and unwanted salts. |
Diagrams
Diagram 1: RNP Degradation Pathways & Stabilization Mechanisms
Diagram 2: Workflow for RNP Formulation & Stability Assessment
Application Notes
Achieving precise tissue and cell selectivity remains a paramount challenge in therapeutic CRISPR-Cas9 RNP delivery. This document details advanced strategies to enhance selectivity through engineered targeting moieties and optimized in vivo administration routes, framed within nanoparticle (NP)-mediated delivery research. The goal is to maximize on-target editing while minimizing off-target effects and immune clearance.
Advanced Targeting Moieties for Nanoparticle Functionalization
Targeting moieties direct NP-RNP complexes to specific cell surface receptors. Selection depends on the target tissue's unique molecular signature (e.g., overexpressed receptors, specific glycans).
- Ligand-Based Targeting: Small molecules (e.g., folate for activated macrophages), peptides (e.g., RGD for integrin αvβ3 on tumor vasculature), and aptamers offer high affinity and modular conjugation.
- Antibody/Fragment-Based Targeting: Monoclonal antibodies (mAbs) or engineered fragments (scFv, Fab) provide exceptional specificity. Humanized or fully human variants reduce immunogenicity.
- Biomimetic Targeting: Coating NPs with cell membranes (e.g., leukocyte, platelet) confers natural homing abilities and immune evasion.
- Conditional Activation: Using "stealth" coatings (PEG) with labile linkers or stimuli-responsive ligands (pH, enzyme-cleavable) that reveal targeting motifs only at the disease site enhances specificity.
Table 1: Comparison of Advanced Targeting Moieties for NP-RNP Delivery
| Moietiy Class | Example (Target Receptor) | Conjugation Method | Typical Editing Enhancement (vs. Non-Targeted) | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Small Molecule | Folate (Folate Receptor) | EDC/NHS, maleimide | 3-5x (in vitro, cancer cells) | Low immunogenicity, stable, cheap | Limited receptor diversity, moderate affinity |
| Peptide | iRGD (αv integrins / NRP-1) | Maleimide, click chemistry | 4-8x (in vivo, tumor tissue) | Good tissue penetration, design flexibility | Proteolytic instability, possible immunogenicity |
| Aptamer | AS1411 (Nucleolin) | Thiol-maleimide, streptavidin-biotin | 5-10x (in vitro, cancer cells) | High affinity, low immunogenicity, chemical stability | Nuclease sensitivity, complex SELEX selection |
| scFv | Anti-EGFR scFv (EGFR) | Genetic fusion to NP coat protein | 10-20x (in vivo, tumor tissue) | Very high specificity and affinity | Potential immunogenicity, complex production |
| Cell Membrane | Leukocyte membrane (ICAM-1) | Membrane extrusion or co-incubation | 6-15x (in vivo, inflamed endothelium) | Innate biocompatibility & homing | Batch-to-batch variability, complex characterization |
2In VivoDelivery Routes and Selectivity Implications
The administration route critically determines first-pass tissue distribution, immune exposure, and ultimate selectivity of NP-RNP complexes.
- Intravenous (IV): The most common route. Selectivity is primarily conferred by NP physicochemical properties (size, charge, surface ligands) and the Enhanced Permeability and Retention (EPR) effect in tumors or leaky vasculature sites. Liver and spleen sequestration is a major hurdle.
- Local/Regional Administration: Direct injection into target tissues (e.g., intratumoral, intramyocardial, intrathecal) maximizes local concentration and minimizes systemic exposure. Often combined with imaging guidance.
- Subcutaneous (SC) / Intramuscular (IM): Primarily for targeting antigen-presenting cells in lymphoid organs for immunotherapeutic applications or for sustained release depots.
- Inhalation (Intranasal / Intratracheal): For direct targeting of lung epithelial cells and airways, bypassing systemic circulation.
Table 2: Quantitative Profile of Key In Vivo Delivery Routes for NP-RNP
| Delivery Route | Typical NP Size Range | Primary Target Tissues/Cells | Approximate Bioavailability (% of dose) | Key Selectivity Mechanism | Major Clearance/Barrier |
|---|---|---|---|---|---|
| Intravenous (IV) | 20-150 nm | Liver (Kupffer cells, hepatocytes), Spleen, Tumors (via EPR) | 2-10% (of injected dose in target tissue) | Passive (EPR) & Active (Ligand-Receptor) | RES/Uptake, Renal Clearance, Protein Corona |
| Intratumoral (IT) | 20-200 nm | Local tumor mass, Tumor-infiltrating immune cells | 40-70% (retained locally) | Physical confinement, local diffusion | Tissue backflow, limited penetration |
| Intracerebroventricular (ICV) | < 50 nm | Brain parenchyma (ependymal, neuronal cells) | 15-30% (distribution in CNS) | Direct bypass of BBB, CSF circulation | CSF turnover, limited parenchymal penetration |
| Inhalation | 10-100 nm | Lung epithelium, Alveolar macrophages | 20-50% (deposition in lungs) | Mucociliary clearance avoidance, alveolar deposition | Macrophage phagocytosis, mucus barrier |
Experimental Protocols
Protocol: Conjugation of scFv Targeting Ligands to Lipid Nanoparticles (LNPs)
Objective: Functionalize CRISPR-Cas9 RNP-loaded LNPs with a single-chain variable fragment (scFv) for cell-specific targeting.
Materials:
- Pre-formed, Cas9 RNP-loaded LNPs with maleimide-terminated PEG-lipids.
- Purified scFv with a C-terminal cysteine residue.
- Reduction buffer: 20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, pH 7.5, with 10 mM TCEP.
- Desalting column (e.g., Zeba Spin, 7K MWCO).
- Nitrogen (N₂) gas stream.
- HEPES Buffered Saline (HBS), pH 7.4.
Procedure:
- scFv Reduction: Incubate scFv (1 mg/mL) in reduction buffer for 1 hour at room temperature to reduce the C-terminal cysteine and generate a free thiol.
- Buffer Exchange: Purify the reduced scFv using a desalting column equilibrated with degassed HBS (pH 7.4) to remove TCEP and exchange buffer. Elute in 0.5 mL HBS.
- Conjugation Reaction: Immediately mix the purified, reduced scFv with maleimide-LNPs at a molar ratio of 50:1 (scFv:maleimide group) in a total volume of 1 mL HBS. Gently flush the headspace with N₂ gas to prevent oxidation.
- Incubation: React for 2 hours at 4°C with gentle end-over-end mixing.
- Purification: Remove unreacted scFv by size exclusion chromatography (e.g., Sepharose CL-4B column) or by dialysis against HBS (100kDa MWCO) overnight at 4°C.
- Characterization: Determine scFv conjugation efficiency via fluorescence assay (if scFv is labeled) or BCA assay on post-purification supernatant. Measure NP size and zeta potential by dynamic light scattering.
Protocol: Evaluating Tropism via Multi-RouteIn VivoAdministration in a Murine Model
Objective: Compare the biodistribution and editing efficiency of untargeted vs. targeted NP-RNPs administered via different routes.
Materials:
- Mice (e.g., C57BL/6, tumor-bearing if applicable).
- Cy5-labeled Cas9 RNP.
- Targeted (scFv-conjugated) and non-targeted LNPs (from Protocol 2.1).
- In vivo imaging system (IVIS) or equivalent.
- Tissue homogenization kit.
- gDNA extraction kit.
- Droplet Digital PCR (ddPCR) or next-generation sequencing (NGS) reagents for indel analysis.
Procedure:
- NP Preparation: Load Cy5-Cas9 RNP into targeted and non-targeted LNPs. Confirm loading and fluorescence.
- Animal Dosing: Divide mice into groups (n=5). Administer a single dose (e.g., 1 mg/kg RNP) via:
- Group A (IV): Tail vein injection, 100 μL volume.
- Group B (IT): Ultrasound-guided injection into tumor, 50 μL volume.
- Group C (Inhalation): Intranasal instillation, 50 μL volume (lightly anesthetized).
- In Vivo Imaging: At 1, 4, 24, and 48 hours post-injection, image mice using IVIS (Cy5 channel) to track whole-body fluorescence distribution.
- Tissue Harvest: At 72 hours, euthanize mice. Harvest target organs (liver, spleen, lung, tumor, etc.) and image ex vivo.
- Genomic Analysis: Homogenize tissues. Extract gDNA. Quantify on-target indel frequency at the target locus using ddPCR (for known edits) or NGS.
- Data Analysis: Correlate fluorescence intensity (biodistribution) with measured editing efficiency in each tissue for each route and NP type.
Diagrams
Diagram 1: Ligand-Targeted NP-RNP Delivery Pathway
Diagram 2: Development Workflow for Targeted Delivery
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Targeted NP-RNP Delivery Research
| Item | Function in Research | Example Product/Catalog |
|---|---|---|
| Maleimide-PEG-DSPE | Provides conjugation handle on NP surface for thiol-linked ligands (peptides, scFv). | Avanti Polar Lipids, 880126P |
| TCEP-HCl | Reduces disulfide bonds to generate free thiols on targeting ligands for conjugation. | Thermo Fisher Scientific, 20490 |
| Zeba Spin Desalting Columns | Rapid buffer exchange to remove reducing agents or unreacted small molecules post-conjugation. | Thermo Fisher Scientific, 89882 |
| Recombinant scFv with Cys-Tag | Engineered targeting ligand with defined site for controlled, oriented conjugation. | Custom from gene synthesis & bacterial expression. |
| Cy5 NHS Ester | Fluorescent dye for labeling Cas9 protein to enable in vivo imaging and biodistribution studies. | Lumiprobe, 23020 |
| Droplet Digital PCR (ddPCR) Supermix | For absolute quantification of low-frequency genome editing events in harvested tissues. | Bio-Rad, 1863024 |
| Sepharose CL-4B | Size exclusion chromatography medium for purifying conjugated NPs from free ligands. | Cytiva, 17015001 |
| In Vivo-JetPEI | A benchmark polymeric transfection agent for comparative studies with novel NP formulations. | Polyplus, 201-50G |
This Application Note provides a framework for transitioning the production of CRISPR-Cas9 Ribonucleoprotein (RNP) complexes loaded into nanoparticle delivery systems from laboratory research and development to clinical-grade Good Manufacturing Practice (GMP) production. The focus is on critical process parameters, quality control, and analytical validation required for Investigational New Drug (IND) application and Phase I clinical trials.
Key Scalability Challenges and Solutions
Table 1: Primary Scalability Challenges in RNP-Nanoparticle Production
| Process Step | Lab-Scale Method | Scale-Up Challenge | Clinical-Grade Solution | Critical Quality Attribute (CQA) Impacted |
|---|---|---|---|---|
| RNP Complex Formation | Manual pipetting, low-concentration assembly. | Batch consistency, RNP complex stability, endotoxin control. | In-line mixing in controlled buffer, UF/DF for buffer exchange, real-time pH/temp monitoring. | RNP bioactivity, endotoxin levels (<5 EU/mg), sub-visible particles. |
| Nanoparticle Formulation | Bulk mixing, manual extrusion through small membranes. | Shear stress denaturation, particle size heterogeneity, aseptic control. | Tangential Flow Filtration (TFF) with controlled shear, sequential microfluidics, sterile single-use flow paths. | Particle Size (PDI <0.2), Zeta Potential, Encapsulation Efficiency (>80%). |
| Purification | Bench-top centrifugation, dialysis. | Low yield, poor separation efficiency, contamination risk. | Scalable Chromatography (e.g., IEX, SEC), automated TFF systems. | Purity (>95%), residual solvent levels, sterility. |
| Sterile Filtration & Fill-Finish | Syringe filtration into vials. | Product adsorption to filter, vial contamination, dose uniformity. | Pre-use filter flushing validation, automated filling in ISO 5 environment, 100% weight check. | Sterility (SAL <10^-6), dose accuracy (±5%), container closure integrity. |
GMP Considerations and Protocol Outline
Master Cell Bank and Recombinant Cas9 Protein Production
Protocol: GMP-Grade Cas9 Protein Expression and Purification
- Starting Material: Use a GMP Master Cell Bank (MCB) of E. coli or HEK293 cells expressing His-tagged Cas9, fully characterized for identity, sterility, and mycoplasma.
- Fermentation/Bioreactor: Scale production in a single-use bioreactor. Monitor and control dissolved oxygen, pH, and temperature.
- Purification: Perform cell lysis, followed by immobilized metal affinity chromatography (IMAC) and size-exclusion chromatography (SEC) in a closed system.
- Formulation & Storage: Diafilter into GMP formulation buffer (e.g., 20 mM HEPES, 150 mM KCl, 10% glycerol, pH 7.5). Perform 0.22 µm sterile filtration. Fill into sterile vials; store at ≤ -60°C.
- Quality Control: Test for identity (SDS-PAGE/Western), purity (HPLC-SEC >98%), potency (in vitro cleavage assay), endotoxin (<5 EU/mg), and bioburden.
GMP-Grade sgRNA Synthesis
Protocol: Large-Scale, HPLC-Purified sgRNA Production
- Synthesis: Use in vitro transcription (IVT) with GMP-grade NTPs and T7 RNA polymerase. Employ DNase treatment.
- Purification: Purify via preparative-scale anion-exchange HPLC. Use only USP Water for Injection (WFI) in buffers.
- Quality Control: Confirm identity (mass spec), purity (denaturing PAGE/Analytical HPLC >90%), integrity (no degradation fragments), endotoxin (<5 EU/mg), and sterility.
Clinical-Grade RNP Complexation and Nanoparticle Loading
Protocol: Scale-Up of Lipid Nanoparticle (LNP) Encapsulation via Microfluidics
- RNP Formation: Dilute GMP Cas9 and sgRNA in complexation buffer (WFI-based). Mix at optimized molar ratio via in-line static mixer. Hold at 2-8°C for 15 min.
- LNP Formulation:
- Prepare an aqueous phase: Formed RNP complex in citrate buffer (pH 4.0).
- Prepare an organic phase: Ionizable lipid (e.g., DLin-MC3-DMA), phospholipid, cholesterol, PEG-lipid in ethanol.
- Use a GMP-compatible microfluidic mixer (e.g., CIJ or SHM) with controlled flow rates (TR 3:1, aqueous:organic) to form LNPs.
- Immediately dilute output in 10x volume of PBS (pH 7.4).
- Buffer Exchange & Concentration: Use TFF with a 100 kDa MWCO Pellicon cassette to exchange into final formulation buffer (e.g., PBS, sucrose) and concentrate to target titer.
- Sterile Filtration: Pass through a 0.22 µm PES sterile filter into a sterile bulk bag.
Table 2: In-Process Controls (IPC) for RNP-LNP Production
| IPC Point | Test Method | Acceptance Criteria | Action if OOS |
|---|---|---|---|
| Post-RNP Mix | Native Gel Shift | >95% complex formation | Adjust ratio, re-mix. |
| Post-Microfluidics | Dynamic Light Scattering | Size: 70-100 nm, PDI <0.15 | Adjust flow rates, filter. |
| Post-TFF | UV-Vis Spectroscopy | [Cas9] within 10% of target | Adjust concentration step. |
| Pre-Fill (Bulk) | pH, Appearance | pH 7.4 ± 0.2, clear to opalescent | Adjust pH, investigate turbidity. |
Analytical Methods and Release Testing
Table 3: Required Release Tests for Clinical-Grade RNP-LNP Drug Product
| Test | Method | Specification |
|---|---|---|
| Identity | Cas9 ELISA, sgRNA Seq | Matches reference standard. |
| Potency | In vitro T7E1 cleavage assay in target cells. | EC50 within ±30% of reference. |
| Purity | RP-HPLC (empty LNPs), Agarose Gel (free RNA). | >85% encapsulation, <5% free sgRNA. |
| Size & PDI | Dynamic Light Scattering (DLS). | 70-100 nm, PDI ≤0.20. |
| Endotoxin | LAL Kinetic Chromogenic Assay. | <5.0 EU/mL. |
| Sterility | USP <71> Membrane Filtration. | No growth after 14 days. |
| Visible/Sub-visible Particles | USP <787>, <788>. | Meets Ph. Eur./USP limits. |
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Materials for Scalable RNP-LNP Process Development
| Item | Function | Example/Supplier Note |
|---|---|---|
| GMP-Grade Cas9 Expression System | Source of active therapeutic protein. | Cell banks must be qualified (COA, TSE/BSE). Use vendor audit reports. |
| Pharmaceutical-Grade Lipids | Formulation of stable, efficacious LNPs. | DLin-MC3-DMA, DSPC, Cholesterol, PEG-DMG. Sourced with GMP DMF. |
| Single-Use Bioreactor & TFF Systems | Scalable, closed-system production and purification. | Sartorius Biostat STR, Repligen TangenX TFF. Minimizes cross-contamination. |
| GMP Microfluidic Mixer | Reproducible, scalable nanoparticle formation. | Precision Nanosystems Ignite or Spark systems with GMP protocols. |
| Process Analytical Technology (PAT) | Real-time monitoring of CPPs (e.g., size, pH). | In-line DLS probes (PSS AccuSizer), pH sensors. Enables QbD. |
| Stability Testing Chambers | ICH-compliant storage condition studies. | Chambers for 25°C/60%RH, 5°C, -20°C, -70°C. |
| Mycoplasma Detection Kit | Essential safety testing for cell-derived products. | PCR-based kits (e.g., Lonza MycoAlert). |
Visualizations
Title: GMP Workflow for RNP-LNP Production
Title: QbD Link Between CPPs and CQAs
Within the development of lipid nanoparticles (LNPs) for CRISPR-Cas9 ribonucleoprotein (RNP) delivery, batch-to-batch variability poses a critical challenge to therapeutic efficacy and safety. This variability can manifest in differences in particle size, encapsulation efficiency, surface charge, and biological activity, directly impacting gene editing outcomes. Establishing robust, multi-parametric quality control (QC) standards is therefore essential for clinical translation. These application notes provide detailed protocols for key characterization assays, framed within the context of LNP-RNP formulation development.
Key Quality Attributes (KQAs) & Analytical Methods
The following table summarizes the critical quality attributes for LNP-RNP complexes and the recommended analytical techniques for their assessment.
Table 1: Key Quality Attributes and Analytical Methods for LNP-RNP Complexes
| Quality Attribute | Target Range/Value | Analytical Method | Impact on Performance |
|---|---|---|---|
| Particle Size & PDI | 70-100 nm, PDI < 0.2 | Dynamic Light Scattering (DLS) | Cellular uptake, biodistribution, potency. |
| Zeta Potential | Slightly negative to neutral (e.g., -10 to +5 mV) | Electrophoretic Light Scattering | Colloidal stability, protein adsorption, cellular interaction. |
| RNP Encapsulation Efficiency (EE%) | > 90% | Ribogreen Fluorescence Assay | Directly correlates with delivered payload and editing efficiency. |
| Total RNP Content | Consistent with theoretical load | BCA/Modified Protein Assay | Dosage accuracy and batch potency. |
| Structural Integrity & Morphology | Spherical, unilamellar vesicles | Cryo-Electron Microscopy (Cryo-EM) | Payload protection and fusion/disruption kinetics. |
| In Vitro Potency (Editing Efficiency) | Batch-specific benchmark (%) | T7E1 Assay or NGS | Functional biological activity; the ultimate efficacy readout. |
| Endotoxin & Sterility | Endotoxin < 1 EU/mL, Sterile | LAL Test, Microbial Culture | Safety for in vivo administration. |
Detailed Experimental Protocols
Protocol 3.1: Determination of RNP Encapsulation Efficiency via Ribogreen Assay
Principle: The Quant-iT Ribogreen reagent exhibits a massive fluorescence enhancement upon binding to RNA. The assay quantifies free (unencapsulated) RNP in the presence of a detergent that lyses LNPs but does not interfere with RNA detection.
Materials:
- LNP-RNP formulation
- Quant-iT Ribogreen RNA Reagent (Thermo Fisher, R11490)
- Tris-EDTA (TE) Buffer, pH 7.5
- Triton X-100 (25% v/v solution)
- Nuclease-free water
- Purified CRISPR-Cas9 RNP standard for calibration curve
- Black-walled, clear-bottom 96-well plate
- Fluorescence microplate reader (Ex: ~480 nm, Em: ~520 nm)
Procedure:
- Prepare Standards: Serially dilute the purified RNP standard in TE buffer to create a calibration curve (e.g., 0-500 ng/mL RNA concentration).
- Prepare Samples:
- Total RNP (Lysed): Dilute LNP-RNP formulation 1:100 in TE buffer containing 0.5% Triton X-100. Incubate for 10 min at RT.
- Free RNP (Unlysed): Dilute LNP-RNP formulation 1:100 in TE buffer only.
- Add Dye: In a 96-well plate, mix 100 µL of each standard or sample with 100 µL of Ribogreen reagent diluted 1:500 in TE buffer. Perform in triplicate.
- Measure Fluorescence: Incubate for 5 min protected from light. Read fluorescence.
- Calculation:
- Determine RNA concentrations from the standard curve.
- EE% = [1 - (Free RNP Concentration / Total RNP Concentration)] x 100.
Protocol 3.2: In Vitro Potency Assessment via T7 Endonuclease I (T7E1) Assay
Principle: This assay detects insertions/deletions (indels) generated by non-homologous end joining after CRISPR-Cas9-mediated DNA cleavage. T7E1 cleaves heteroduplex DNA formed by annealing wild-type and indel-containing PCR products.
Materials:
- Cells treated with LNP-RNP (e.g., HEK293 containing a GFP reporter)
- Genomic DNA extraction kit
- PCR primers flanking the target site
- High-fidelity PCR Master Mix
- T7 Endonuclease I (NEB, M0302S)
- NEBuffer 2.1
- Agarose gel electrophoresis system
Procedure:
- Harvest & Extract gDNA: 72 hours post-transfection, harvest cells and extract genomic DNA.
- Amplify Target Locus: Perform PCR (~300-500 bp product) using target-specific primers.
- Heteroduplex Formation: Denature PCR products at 95°C for 5 min, then re-anneal by ramping down to 25°C at -0.1°C/sec.
- T7E1 Digestion: Digest re-annealed products with T7E1 enzyme (1 µL per 20 µL reaction) for 60 min at 37°C.
- Analysis: Run digested products on a 2% agarose gel. Cleavage fragments indicate presence of indels.
- Quantification: Use gel image analysis software (e.g., ImageJ) to measure band intensities.
- Indel % = 100 x [1 - sqrt(1 - (b + c)/(a + b + c))], where a is the intensity of the undigested band, and b & c are the cleavage products.
Visualizations
- Diagram 1 Title: LNP-RNP QC and Batch Release Workflow
- Diagram 2 Title: LNP-RNP Delivery Pathway & Variability Impact Points
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 2: Key Reagent Solutions for LNP-RNP QC and Potency Assays
| Item | Supplier/Example | Function in Context |
|---|---|---|
| Ionizable Lipid (e.g., DLin-MC3-DMA) | MedKoo, Avanti | Critical structural lipid for LNP formation and endosomal escape; primary source of batch variability. |
| Quant-iT Ribogreen RNA Reagent | Thermo Fisher (R11490) | Fluorescent dye for sensitive, specific quantification of encapsulated vs. free RNA/RNP. |
| T7 Endonuclease I | New England Biolabs (M0302S) | Enzyme for detecting CRISPR-Cas9-induced indels in genomic DNA via mismatch cleavage. |
| Microfluidic Device (NanoAssemblr) | Precision NanoSystems | Enables reproducible, scalable LNP formulation with controlled mixing kinetics. |
| Size & Zeta Reference Standards | Malvern Panalytical | Polystyrene beads of known size and zeta potential for instrument calibration and validation. |
| Recombinant CRISPR-Cas9 Protein | IDT, Thermo Fisher | High-purity, endotoxin-free Cas9 protein for RNP complex assembly; critical for batch consistency. |
| Endotoxin Detection Kit (LAL) | Lonza, Thermo Fisher | For safety testing to ensure LNP preparations meet injectable standards (<1 EU/mL). |
Benchmarking Success: Analytical Methods, Model Systems, and Platform Comparisons
Within a broader thesis investigating CRISPR-Cas9 ribonucleoprotein (RNP) delivery via engineered nanoparticles, the precise quantification of genome editing outcomes is paramount. This application note details three primary techniques—Next-Generation Sequencing (NGS), T7 Endonuclease I (T7E1) assay, and Tracking of Indels by Decomposition (TIDE)—for evaluating editing efficiency and characterizing the spectrum of insertion and deletion (indel) mutations following nanoparticle-mediated RNP delivery. Each method offers distinct advantages in throughput, resolution, and cost.
Comparative Analysis of Quantification Methods
The following table summarizes the key characteristics, advantages, and limitations of NGS, T7E1, and TIDE.
Table 1: Comparison of Genome Editing Quantification Methods
| Method | Throughput | Resolution | Quantitative Output | Primary Advantage | Key Limitation | Approximate Cost per Sample |
|---|---|---|---|---|---|---|
| NGS (Amplicon-Seq) | High (Multiplexed) | Nucleotide-level | Percentage and exact sequence of each indel | Comprehensive, unbiased characterization of all mutation types; high sensitivity. | High cost, complex data analysis, longer turnaround time. | $50 - $150 |
| T7E1 Assay | Low to Medium | Detection of heteroduplexes | Estimated total indel percentage (%) | Rapid, low-cost, equipment accessible. | Low sensitivity (<~5%), no indel sequence info, prone to false positives/negatives. | $5 - $20 |
| TIDE Analysis | Medium | Decomposition of indel profiles up to ~20 bp | Percentage of major indels and total editing efficiency (%) | Rapid, provides basic indel profile from Sanger sequencing. | Limited resolution of complex mixtures, relies on Sanger sequencing quality. | $15 - $40 (seq cost) |
Detailed Protocols
Protocol 1: T7 Endonuclease I (T7E1) Mismatch Cleavage Assay
Application: Rapid, semi-quantitative estimation of editing efficiency post nanoparticle-RNP transfection.
Materials:
- Genomic DNA extracted from treated/control cells.
- PCR primers flanking the target site.
- High-fidelity PCR master mix.
- T7 Endonuclease I (commercially available).
- NEBuffer 2 (or buffer supplied with enzyme).
- Agarose gel electrophoresis system.
Procedure:
- PCR Amplification: Amplify the target genomic region (200-500 bp) from ~100 ng gDNA using a high-fidelity polymerase. Include a control sample from untreated cells.
- Heteroduplex Formation: Purify PCR products. Denature and reanneal to form heteroduplexes: 95°C for 10 min, ramp down to 85°C at -2°C/s, then to 25°C at -0.1°C/s.
- T7E1 Digestion: Prepare a 20 µL reaction containing 200-400 ng reannealed PCR product, 1X NEBuffer 2, and 1 µL (5-10 units) of T7 Endonuclease I. Incubate at 37°C for 30-60 minutes.
- Analysis: Run digested products on a 2-2.5% agarose gel. Cleaved fragments indicate presence of indels.
- Quantification: Calculate approximate indel frequency using densitometry: % Indels = 100 × [1 - sqrt(1 - (b+c)/(a+b+c))], where a is integrated intensity of undigested band, and b & c are digested fragment intensities.
Protocol 2: Tracking of Indels by Decomposition (TIDE) Analysis
Application: Rapid quantification of editing efficiency and decomposition of major indel patterns from Sanger sequencing data.
Materials:
- Genomic DNA from edited and control populations.
- Sanger sequencing service/capability.
- TIDE web tool (https://tide.nki.nl) or similar software (ICE, Synthego).
Procedure:
- PCR and Sequencing: Amplify the target region from test and control samples using a primer ~100-200 bp from the cut site for sequencing. Purify PCR product and submit for Sanger sequencing with the same primer.
- Data Preparation: Obtain .ab1 chromatogram files for both control (reference) and edited samples.
- TIDE Analysis:
- Navigate to the TIDE web interface.
- Upload the control sample .ab1 file as the "Reference trace."
- Upload the edited sample .ab1 file as the "Edited trace."
- Define the target sequence and the expected cut site position relative to the sequencing trace.
- Set the decomposition window (typically ~15 bp on each side of the cut site) and the maximum indel size to search (e.g., ±20 bp).
- Run the decomposition analysis.
- Interpretation: The tool outputs the overall editing efficiency (%) and a detailed breakdown of the most frequent indel sequences and their individual percentages. The R² value indicates quality of fit.
Protocol 3: Next-Generation Sequencing (NGS) for Editing Analysis
Application: Gold-standard, high-resolution quantification of all mutagenic outcomes.
Materials:
- Genomic DNA.
- Tailored primers with Illumina adapter overhangs.
- High-fidelity PCR master mix.
- PCR clean-up kit.
- Indexing primers (i7, i5).
- Access to NGS platform (e.g., MiSeq).
Procedure:
- Amplicon Library Preparation:
- Perform a first-round PCR to amplify the target locus from ~50 ng gDNA using primers containing partial Illumina adapter sequences.
- Clean up PCR products.
- Perform a second, limited-cycle PCR to add full dual indices (i7, i5) and flow cell binding sequences.
- Pool and purify the final libraries. Quantify by qPCR or bioanalyzer.
- Sequencing: Dilute library to appropriate concentration and sequence on a MiSeq or similar platform using a paired-end 2x250 or 2x300 cycle kit to ensure overlap across the target site.
- Bioinformatic Analysis:
- Demultiplex reads by sample indices.
- Merge paired-end reads.
- Align reads to the reference amplicon sequence.
- Quantify reads with perfect matches versus those containing indels or substitutions around the target site using tools like CRISPResso2, AmpliCan, or custom Python/R scripts.
- Output: Generate a table and plots detailing total indel percentage, spectrum and frequency of each specific indel, and allele frequencies.
Diagrams
Title: Workflow for Quantifying CRISPR Editing Post-Nanoparticle Delivery
Title: T7E1 Assay Principle and Steps
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Materials for Editing Quantification
| Item | Function/Application | Example Vendor/Product |
|---|---|---|
| High-Fidelity DNA Polymerase | Accurate amplification of target locus from gDNA for NGS, TIDE, or T7E1 input. | NEB Q5, Thermo Fisher Phusion. |
| T7 Endonuclease I | Enzyme that cleaves mismatches in DNA heteroduplexes for the T7E1 assay. | NEB M0302S, IDT. |
| Genomic DNA Extraction Kit | Reliable isolation of high-quality gDNA from transfected cell populations. | Qiagen DNeasy Blood & Tissue, Zymo Quick-DNA. |
| PCR Purification Kit | Clean-up of amplicons prior to sequencing, T7E1, or further processing. | Zymo DNA Clean & Concentrator, Qiagen MinElute. |
| Illumina-Compatible Indexing Primers | For multiplexed sample preparation in NGS amplicon sequencing. | Illumina Nextera XT, IDT for Illumina. |
| Sanger Sequencing Service | Generation of chromatogram files for TIDE analysis. | Genewiz, Eurofins, in-house facility. |
| CRISPResso2 Software | Standardized, open-source bioinformatics pipeline for analyzing NGS data from CRISPR experiments. | Open source (GitHub). |
| Nucleofection System/Reagent | (Control Transfection) Positive control for nanoparticle-RNP delivery optimization. | Lonza Nucleofector, Lipofectamine CRISPRMAX. |
| Synthetic Indel Standard | Control DNA with known indel mixtures for assay validation and standardization. | Custom gBlocks from IDT. |
Within the broader thesis on CRISPR-Cas9 ribonucleoprotein (RNP) delivery via nanoparticles, a critical component is the comprehensive assessment of off-target editing. Nanoparticle delivery can influence RNP kinetics and persistence, potentially altering the off-target profile compared to plasmid-based delivery. This application note details three cornerstone methodologies for off-target assessment: experimental techniques GUIDE-seq and CIRCLE-seq, and computational in silico prediction tools, providing protocols framed within an RNP context.
Table 1: Comparative Summary of Off-Target Assessment Methods
| Method | Principle | Detection Sensitivity | Throughput | Requires Pre-knowledge of Sites? | Key Advantage | Key Limitation |
|---|---|---|---|---|---|---|
| GUIDE-seq | Capture of double-stranded oligodeoxynucleotide tags into double-strand breaks (DSBs) in situ. | ~0.1% of indels (in cells) | Medium | No | Identifies off-targets in relevant cellular context with chromatin. | Lower sensitivity than in vitro methods; requires tag delivery. |
| CIRCLE-seq | In vitro circularization and amplification of sheared genomic DNA, followed by Cas9 RNP cleavage and sequencing. | ~0.01% of reads (in vitro) | High | No | Ultra-sensitive, genome-wide, cell-context independent. | Does not reflect intracellular chromatin or delivery conditions. |
| In Silico Prediction | Algorithmic scoring based on sequence similarity to on-target and chromatin accessibility data. | Not applicable (predictive) | Very High | Yes (for sequence-based scoring) | Fast, inexpensive, guides experimental design. | Prone to false positives and negatives; depends on algorithm. |
Table 2: Typical Experimental Output Metrics (Representative Data)
| Method | Metric | Typical Range for a Well-Designed sgRNA (RNP Delivery) | Notes for Nanoparticle-Delivered RNP |
|---|---|---|---|
| GUIDE-seq | Number of identified off-target sites | 0 - 20 sites | Can be influenced by nanoparticle unpacking rate and RNP durability. |
| GUIDE-seq | Read ratio (Off-target / On-target) | 0.001% - 10% | Reflects relative editing efficiency in cellular context. |
| CIRCLE-seq | Number of identified off-target sites | 10 - 100+ sites | Represents maximum potential off-target landscape; numbers may be higher than GUIDE-seq. |
| CIRCLE-seq | Cleavage score / frequency | Varies widely by site | Purely sequence-dependent; not modulated by cellular delivery. |
| In Silico | Top predicted off-target sites | 1 - 50 sites (commonly reviewed) | Should be validated experimentally; chromatin data improves predictions. |
Detailed Experimental Protocols
Protocol 1: GUIDE-seq for Nanoparticle-Delivered RNP
Adapted from Tsai et al., Nat Biotechnol, 2015, for RNP contexts.
I. Key Research Reagent Solutions
| Reagent / Material | Function in Protocol |
|---|---|
| Cas9 Nuclease (purified) & sgRNA (chemically modified) | Form the pre-complexed RNP for delivery. |
| GUIDE-seq Oligoduplex (dsODN) | Double-stranded tag that integrates into DSBs. Critical for off-target capture. |
| Polymerase-based Nanoparticles (e.g., PGNP) | Delivery vehicle for co-delivering RNP and dsODN. |
| PCR Reagents for GUIDE-seq (Primers, Polymerase) | Amplify genomic regions flanking integrated dsODN tags. |
| High-Throughput Sequencing Platform | For final library sequencing and off-target identification. |
II. Step-by-Step Workflow
- Complex Formation: Pre-complex purified Cas9 protein with synthetic, chemically modified sgRNA to form RNP. Anneal GUIDE-seq dsODN tag separately.
- Nanoparticle Formulation: Co-load or co-formulate the RNP and dsODN tag into your selected nanoparticle system (e.g., polymeric, lipid-based). Determine optimal N:P ratio and encapsulation efficiency.
- Cell Delivery & Editing: Deliver nanoparticle formulation to target cells (e.g., HEK293T, primary T cells). Include controls (RNP only, dsODN only). Incubate for 48-72 hours.
- Genomic DNA Extraction: Harvest cells and extract high-molecular-weight gDNA using a silica-membrane column.
- dsODN Integration Enrichment: Perform a first PCR (~15 cycles) using a primer specific to the dsODN tag and a primer binding to a common adapter ligated to sheared gDNA. This enriches fragments containing the tag.
- Library Amplification & Barcoding: Perform a second, nested PCR to add Illumina sequencing adapters and sample barcodes.
- Sequencing & Analysis: Pool libraries and sequence on a MiSeq or HiSeq platform. Analyze reads using the GUIDE-seq analysis software (available on GitHub) to map tag integration sites and identify off-target loci.
Protocol 2: CIRCLE-seq forIn VitroOff-Target Profiling
Adapted from Tsai et al., Nat Methods, 2017.
I. Key Research Reagent Solutions
| Reagent / Material | Function in Protocol |
|---|---|
| Purified Genomic DNA | Substrate for in vitro cleavage. Isolate from target cell type. |
| Cas9 Nuclease (purified) & sgRNA | The RNP complex for in vitro cleavage reaction. |
| Circligase ssDNA Ligase | Enzymatically circularizes sheared, end-repaired genomic DNA fragments. |
| Phi29 Polymerase | Performs rolling circle amplification of circularized DNA. |
| T7 Endonuclease I or Surveyor Nuclease | Detects cleavage-induced mismatches in re-annealed amplicons (optional validation step). |
II. Step-by-Step Workflow
- Genomic DNA Isolation & Shearing: Extract gDNA from the relevant cell type. Mechanically shear to ~300 bp fragments using a Covaris sonicator.
- End-Repair & Circularization: Repair fragment ends using a polishing enzyme mix. Purify DNA and perform in vitro circularization using Circligase ssDNA ligase.
- Rolling Circle Amplification (RCA): Treat circularized DNA with exonuclease to linearize any non-circular DNA. Amplify circular templates using phi29 polymerase via RCA.
- In Vitro Cleavage: Incubate the RCA-amplified product with pre-assembled Cas9 RNP (using the same sgRNA as therapeutic candidate). This cleaves sites present in the DNA library.
- Library Preparation for Sequencing: Repair the ends of the cleaved fragments, ligate sequencing adapters, and perform a final PCR.
- Sequencing & Analysis: Sequence the library to high depth. Use the CIRCLE-seq analysis pipeline to identify cleavage sites by detecting fragment ends that map to the genome, signifying RNP cleavage events.
Protocol 3: IntegratingIn SilicoPredictions
Workflow: Use a combination of algorithms to generate a candidate list for experimental validation.
- Primary Sequence-Based Prediction: Input your 20-23 nt sgRNA sequence into tools like CRISPOR (crispor.tefor.net) or CHOPCHOP. These rank potential off-target sites by sequence similarity (allowing mismatches, bulges).
- Integrate Chromatin Context: For predictions relevant to your target cell type, use tools like CROP-IT or input cell-specific ATAC-seq or DNase-seq data (available from ENCODE) to filter or re-rank predictions based on chromatin accessibility.
- Cross-Reference & Prioritize: Compile a union list of top predictions from multiple tools. Prioritize sites within exonic regions or regulatory elements for downstream validation.
- Experimental Validation: Validate top-ranked in silico sites, and all sites from GUIDE-seq/CIRCLE-seq, using targeted deep sequencing (amplicon-seq) of genomic DNA from nanoparticle-treated cells.
Visualized Workflows and Relationships
Title: Integrated Workflow for Off-Target Assessment in RNP Research
Title: Experimental Workflows of CIRCLE-seq vs. GUIDE-seq
CRISPR-Cas9 ribonucleoprotein (RNP) delivery via nanoparticles represents a transformative approach for precise genome editing. Its success is critically dependent on the choice of in vitro model system. Primary cells offer physiological relevance but are fragile and limited in supply. Stem cells, including induced pluripotent stem cells (iPSCs), provide scalability and differentiation potential but require careful maintenance of pluripotency or directed differentiation. Hard-to-transfect cell lines, such as Jurkat T cells or primary neurons, present significant barriers to conventional transfection methods. Nanoparticle-mediated RNP delivery must be optimized for each system to achieve high editing efficiency while maintaining cell viability and function.
Comparative Analysis of Model Systems for CRISPR RNP Delivery
Table 1: Characteristics and CRISPR RNP Delivery Challenges of In Vitro Model Systems
| Model System | Key Characteristics | Advantages for Genome Editing | Challenges for NP-RNP Delivery | Typical Editing Efficiency Range* |
|---|---|---|---|---|
| Primary Cells (e.g., PBMCs, fibroblasts) | Non-immortalized, finite lifespan, high physiological relevance. | Disease modeling, autologous therapies, avoids cell line artifacts. | Low proliferation, sensitive to manipulation, donor variability, limited material. | 10-40% (highly variable by cell type) |
| Stem Cells (e.g., iPSCs, hESCs) | Self-renewal, pluripotent, can differentiate into any cell type. | Isogenic cell line generation, developmental studies, scalable source. | Difficulty in transfection, need to retain pluripotency post-editing, clonal selection required. | 20-70% (depends on passage and status) |
| Hard-to-Transfect Cell Lines (e.g., Jurkat, THP-1, Neurons) | Often suspension, non-adherent, or post-mitotic; complex morphology. | Models for immunology, neurobiology, cancer. | Low endocytic uptake, membrane composition, toxicity sensitivity. | 15-60% (requires optimized NPs) |
*Efficiency is highly dependent on nanoparticle formulation, target locus, and cell state.
Table 2: Nanoparticle Formulation Strategies for Different Cell Models
| Cell Model | Recommended NP Type | Key Functionalization | Rationale | Critical Optimization Parameter |
|---|---|---|---|---|
| Primary Human T Cells | Biodegradable polymeric NPs (e.g., PLGA) | CD3 or CD28 antibodies (surface conjugation) | Enhances targeting and uptake via receptor-mediated endocytosis. | NP size (<150 nm), charge (near-neutral), antibody density. |
| iPSCs | Lipid Nanoparticles (LNPs) | Integrin-targeting peptides (e.g., RGD) | Exploits active adhesion pathways in pluripotent cells. | Molar ratio of ionizable lipid:helper lipid:peptide-lipid. |
| Jurkat T Cells | Cationic Polymer-based NPs (e.g., PBAE) | Poly(ethylene glycol) (PEG) shell | Balances cationic charge for RNP complexation with reduced cytotoxicity. | N:P ratio (polymer nitrogen to RNP phosphate). |
| Primary Neurons | Gold Nanoparticles (AuNPs) | Neuron-penetrating peptides (e.g., RVG) | Facilitates crossing of complex cellular membranes. | Peptide conformation on AuNP surface, RNP loading method. |
Application Notes & Protocols
Protocol 1: CRISPR RNP Delivery to Primary Human Peripheral Blood Mononuclear Cells (PBMCs) Using Targeted Polymeric Nanoparticles
Objective: To achieve knockout of PDCD1 (PD-1) in primary human T cells.
Materials (Research Reagent Solutions):
- PLGA-PEG-Maleimide Nanoparticles: Biodegradable core for RNP encapsulation and stable circulation.
- Anti-CD3e Antibody (Fab' fragment): For targeted delivery to T cells via the TCR complex.
- CRISPR-Cas9 RNP: Pre-complexed Alt-R S.p. Cas9 Nuclease V3 and synthetic sgRNA targeting PDCD1.
- X-tremeGENE HP Transfection Reagent (Control): Commercial reagent for comparison.
- Cell Activation Cocktail: Anti-CD3/CD28 beads to stimulate T cell proliferation post-editing.
Method:
- NP Functionalization: Thiolated anti-CD3e Fab' fragments are conjugated to maleimide groups on PLGA-PEG-NPs via Michael addition. Unconjugated antibody is removed by centrifugation (50,000 x g, 30 min).
- RNP Encapsulation: Cas9 RNP complexes are loaded into NPs using a double emulsion solvent evaporation technique. Briefly, an aqueous RNP solution is emulsified in dichloromethane containing dissolved polymer, then emulsified again in a PVA solution. Solvent is evaporated overnight.
- PBMC Isolation & Activation: Isolate PBMCs from leukapheresis product using Ficoll density gradient centrifugation. Activate T cells using anti-CD3/CD28 beads (1 bead:2 cells) in RPMI-1640 + 10% FBS + 100 IU/mL IL-2 for 48 hours.
- Transfection: Wash activated cells. Resuspend 1e6 cells in 100 µL of serum-free Opti-MEM. Add NP-RNP formulations (equivalent to 5 µg Cas9). For control, use 2 µL X-tremeGENE with 1 µg RNP. Incubate 4-6 hours at 37°C.
- Recovery & Analysis: Replace with complete media + IL-2. After 72 hours, analyze cells by flow cytometry for PD-1 surface expression (knockout efficiency) and Annexin V/propidium iodide staining (viability). Extract genomic DNA for T7 Endonuclease I assay or next-generation sequencing (NGS) of the target locus.
Protocol 2: Gene Editing in Human iPSCs Using Peptide-Modified Lipid Nanoparticles
Objective: To correct a point mutation in BRCA1 via HDR in human iPSCs.
Materials (Research Reagent Solutions):
- Ionizable Lipidoid-based LNP (e.g., C12-200): Enables efficient encapsulation and endosomal escape of RNP and ssODN donor.
- RGD-PEG-DSPE Lipid: Integrin-targeting ligand conjugated to a lipid anchor for incorporation into LNP membrane.
- Cas9 RNP + ssODN Donor Template: RNP complex and a 200 nt single-stranded oligodeoxynucleotide donor with homology arms and the corrective sequence.
- mTeSR1 Medium: Defined, feeder-free culture medium for maintaining pluripotency.
- CloneR Supplement: Enhances survival of single iPSCs post-transfection for clonal expansion.
Method:
- LNP Formulation: LNPs are formulated via rapid microfluidic mixing. The aqueous phase contains RNP and ssODN in citrate buffer (pH 4.0). The organic phase contains ionizable lipidoid, cholesterol, DSPC, and RGD-PEG-DSPE in ethanol. Mix at a 3:1 aqueous:organic flow rate ratio.
- iPSC Culture: Maintain iPSCs in mTeSR1 on Matrigel-coated plates. Passage as clumps using EDTA.
- Transfection: Prior to transfection, dissociate iPSCs to single cells using Accutase. Seed 2e5 cells/well in a 24-well plate in mTeSR1 + CloneR. After 24 hours, add LNP formulations (Cas9 dose: 2 µg/well). Incubate for 48 hours.
- Clonal Isolation & Screening: Transfer cells to a 10 cm dish at low density. After 7-10 days, pick individual colonies using cloning discs. Expand each clone in a 96-well plate. Screen genomic DNA by PCR and Sanger sequencing for precise HDR correction. Confirm pluripotency marker (OCT4, NANOG) expression via immunocytochemistry in corrected clones.
The Scientist's Toolkit: Key Reagents for NP-Mediated RNP Delivery
Table 3: Essential Research Reagent Solutions
| Item | Function in NP-RNP Delivery | Example Product/Brand |
|---|---|---|
| Recombinant High-Fidelity Cas9 Protein | CRISPR nuclease component of the RNP; must be pure, endotoxin-free, and have high specificity. | Alt-R S.p. Cas9 Nuclease V3 (IDT), TruCut Cas9 Protein (Thermo) |
| Chemically Modified Synthetic sgRNA | Guides Cas9 to target DNA; chemical modifications (e.g., 2'-O-methyl, phosphorothioate) enhance stability. | Alt-R CRISPR-Cas9 sgRNA (IDT), Synthego sgRNA |
| Ionizable/Cationic Lipids or Polymers | Core component of NPs that complexes/encapsulates RNP and mediates endosomal escape. | C12-200 lipidoid, Poly(beta-amino ester)s (PBAEs), JetOptimus (Polyplus) |
| Targeting Ligands | Antibodies, peptides, or aptamers conjugated to NP surface for cell-specific delivery. | Anti-CD3 Fab', RGD peptide, Cholesterol-conjugated aptamers |
| HDR Donor Template | Single-stranded oligodeoxynucleotide (ssODN) or double-stranded DNA (dsDNA) for precise edits. | Ultramer DNA Oligos (IDT), gBlocks (IDT) |
| Cell Health/Viability Assay | Measures cytotoxicity of NP formulations (critical for sensitive primary cells). CellTiter-Glo, Annexin V/Flow Kit | |
| High-Sensitivity Genome Editing Analysis | Quantifies indel or HDR efficiency at the target locus. | TIDE assay, Illumina MiSeq for NGS, droplet digital PCR (ddPCR) |
Visualizations
Title: Workflow for Primary T Cell Editing with Targeted NPs
Title: Mechanism of NP-Mediated RNP Delivery and Endosomal Escape
Application Notes
Within the broader research thesis on CRISPR-Cas9 ribonucleoprotein (RNP) delivery via nanoparticles, in vivo validation in murine models is the critical translational step. It assesses the therapeutic potential, biodistribution, efficacy, and pharmacodynamics of the nanoparticle-RNP complex. This document outlines core protocols and readouts for validating gene editing therapeutics in mice.
The primary objectives are: 1) To demonstrate targeted gene modification in relevant tissues, 2) To quantify editing efficacy and correlate it with nanoparticle pharmacokinetics, 3) To measure phenotypic correction or disease-modifying effects, and 4) To evaluate initial safety and off-target effects. Successful validation requires careful selection of the murine model (e.g., wild-type, transgenic, knockout, or humanized), administration route (e.g., intravenous, local injection), and temporal analysis of readouts.
Key Efficacy and Pharmacodynamics Readouts
Efficacy is measured by the functional consequence of editing, while pharmacodynamics (PD) describes the relationship between drug concentration (nanoparticle/RNP) and its pharmacological effect (editing). PD lags behind pharmacokinetics (PK) and must be tracked over time.
Table 1: Core In Vivo Efficacy and Pharmacodynamics Readouts
| Readout Category | Specific Assay | Quantitative Output | Typical Timepoint Post-Injection |
|---|---|---|---|
| Gene Editing | NGS of target locus | Indel frequency (%), precise edit rate (%) | 3-7 days (peak), up to months |
| Target Protein Modulation | Western Blot, ELISA | Protein reduction/expression (%) | 1-4 weeks |
| Transcriptional Effect | qRT-PCR | mRNA level change (fold-change) | 3-14 days |
| Phenotypic Correction | Disease-specific assay (e.g., serum metabolite, imaging) | Normalization of pathological metric | 1 week - several months |
| Biodistribution (PD Correlation) | Bioluminescence/fluorescence imaging, qPCR of gDNA | Signal intensity or RNP vector copies per µg DNA | 6 hrs - 7 days |
Detailed Protocols
Protocol 1: Systemic Delivery and Tissue Harvest for Editing Analysis
Objective: To administer nanoparticle-RNP complexes intravenously and collect tissues for molecular analysis of editing.
Materials: Purified Cas9 protein, sgRNA, formulated nanoparticles, sterile PBS, syringes (29G), C57BL/6 mice (or disease model), dissection tools, DNA/RNA stabilization buffer.
Procedure:
- RNP Complex Formation: Incubate Cas9 protein with sgRNA at a 1:1.2 molar ratio in nuclease-free buffer for 10 min at 25°C.
- Nanoparticle Formulation: Mix pre-formed RNP complex with cationic or lipid nanoparticles per optimized encapsulation protocol. Characterize size and charge (e.g., ~100 nm, slightly negative zeta potential).
- IV Injection: Inject mice via tail vein with a single dose (e.g., 5 mg/kg nanoparticle dose, containing 1-2 µg RNP). Control groups receive PBS or empty nanoparticles.
- Tissue Harvest: Euthanize mice at predetermined timepoints (e.g., 3, 7, 14, 28 days). Harvest target organs (liver, spleen, lung, etc.) and disease-relevant tissues. Snap-freeze in liquid N₂ or store in stabilization buffer.
- Genomic DNA Extraction: Homogenize tissue, extract high-molecular-weight gDNA using a commercial kit. Quantify DNA concentration.
Protocol 2: Analysis of Editing Efficiency via Next-Generation Sequencing (NGS)
Objective: To quantify indel frequencies at the on-target locus from harvested tissues.
Procedure:
- PCR Amplification: Design primers flanking the target site (amplicon ~300bp). Perform PCR on 100 ng of tissue gDNA using a high-fidelity polymerase.
- Library Preparation: Clean PCR amplicons and attach dual-index barcodes using a commercial library prep kit. Pool equimolar amounts from each sample.
- NGS Sequencing: Run on an Illumina MiSeq (2x250 bp) to achieve >10,000x coverage per sample.
- Data Analysis: Use CRISPR-specific analysis pipelines (e.g., CRISPResso2). Align reads to reference sequence and quantify percentage of reads containing insertions or deletions.
Table 2: Example NGS Editing Data from Murine Liver (7 Days Post-IV Dose)
| Nanoparticle Formulation | Dose (mg/kg) | Mean Indel % (Liver) | SD | Predominant Indel Type |
|---|---|---|---|---|
| Lipid Nanoparticle (LNP) A | 5.0 | 45.2% | ±3.5 | -1 bp deletion |
| Polymer Nanoparticle B | 5.0 | 22.7% | ±2.8 | +1 bp insertion |
| PBS Control | - | 0.05% | ±0.02 | N/A |
Protocol 3: Functional Phenotypic Readout – Serum Protein Reduction
Objective: To measure knockdown of a target hepatocyte-secreted protein (e.g., PCSK9, Transthyretin) as a pharmacodynamic biomarker.
Procedure:
- Blood Collection: Perform retro-orbital or submandibular bleeds at baseline and weekly post-injection. Centrifuge to isolate serum.
- ELISA: Use a commercial mouse-specific ELISA kit for the target protein. Run samples in duplicate.
- Analysis: Calculate serum concentration from standard curve. Express as percentage reduction from pre-dose baseline.
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions for Murine RNP Validation
| Reagent/Material | Function & Importance | Example Vendor/Catalog |
|---|---|---|
| Recombinant S. pyogenes Cas9 Protein | Nuclease component of RNP; high purity ensures activity and minimizes immunogenicity. | IDT, Thermo Fisher Scientific |
| Chemically Modified sgRNA | Guides Cas9 to target DNA; chemical modifications (2'-O-methyl, phosphorothioate) enhance stability in vivo. | Synthego, Trilink Biotechnologies |
| Ionizable Cationic Lipid (e.g., DLin-MC3-DMA) | Key component of LNPs for encapsulating RNP and enabling endosomal escape in hepatocytes. | Avanti Polar Lipids, MedChemExpress |
| In Vivo-JetPEI | A polymeric nanoparticle transfection reagent for local or systemic delivery. | Polyplus-transfection |
| Luciferase Reporter Mouse Model | Enables real-time, non-invasive tracking of editing/expression via bioluminescence imaging. | The Jackson Laboratory |
| CRISPResso2 Software | Critical bioinformatics tool for precise quantification of NGS editing outcomes from complex tissue samples. | Open source |
Visualizations
Title: In Vivo Workflow for Systemic Nanoparticle-RNP Delivery
Title: LNP-RNP Delivery and Liver Cell Uptake Pathway
1. Introduction and Context This application note provides a structured comparison of three dominant nanoparticle (NP) platforms—Lipid Nanoparticles (LNPs), Polymeric Nanoparticles (PNPs), and Gold Nanoparticles (AuNPs)—for the intracellular delivery of CRISPR-Cas9 ribonucleoprotein (RNP) complexes. Efficient, transient, and non-viral RNP delivery remains a critical hurdle in therapeutic genome editing. This analysis, framed within a thesis on RNP delivery, evaluates these vectors on the core parameters of delivery efficiency, cytotoxicity, and payload capacity to guide selection for specific research and development applications.
2. Quantitative Comparative Summary
Table 1: Core Performance Metrics for RNP Delivery
| Parameter | Lipid Nanoparticles (LNPs) | Polymeric Nanoparticles (e.g., PEI) | Gold Nanoparticles (AuNPs) |
|---|---|---|---|
| Typical Size Range | 70-120 nm | 50-200 nm | 10-100 nm |
| Encapsulation Efficiency (RNP) | High (60-90%) | Moderate to High (40-85%) | Low to Moderate (via surface conjugation) |
| Payload Capacity | High (Large cargo volume) | High (Tunable polymer matrix) | Low (Surface area-limited) |
| Delivery Efficiency (Cellular Uptake) | Very High (Endocytosis/fusion) | High (Proton-sponge effect) | Moderate (Receptor-mediated) |
| Endosomal Escape | Excellent (Ionizable lipids) | Excellent (Proton sponge) | Poor (Often requires adjuncts) |
| In Vitro Toxicity | Low to Moderate (Dose-dependent) | High (Cationic charge-induced) | Low (With good capping) |
| In Vivo Clearance | Rapid (RES, liver tropic) | Variable (Can be slow) | Slow (Potential long-term retention) |
| Immunogenicity | Moderate (Component-driven) | High (e.g., PEI) | Low (With PEGylation) |
| Manufacturing Scalability | High (Established for mRNA) | Moderate | High |
| Typical RNP Loading Method | Aqueous encapsulation during formation | Complexation/encapsulation | Covalent/Non-covalent surface attachment |
Table 2: Exemplary In Vitro Editing Data (HeLa Cells)
| NP Platform | Formulation Example | Editing Efficiency (% INDELs) | Cell Viability (%) | Key Citation Insight |
|---|---|---|---|---|
| LNP | Ionizable lipid (DLin-MC3-DMA), PEG-lipid | 45-75% | 70-85% | Co-encapsulation of sgRNA and Cas9 protein is optimal. |
| Polymer | Polyethyleneimine (PEI, 25kDa) | 30-60% | 40-65% | High efficiency offset by significant cytotoxicity. |
| Gold NP | 15nm AuNP, PEGylated, RNP conjugated | 20-40% | >90% | Editing efficiency is highly dependent on RNP surface orientation. |
3. Detailed Experimental Protocols
Protocol 3.1: Formulation of Ionizable LNP for RNP Encapsulation Objective: Prepare LNPs encapsulating Cas9 RNP using microfluidic mixing. Materials: Ionizable lipid (e.g., DLin-MC3-DMA), phospholipid (DSPC), cholesterol, PEG-lipid (DMG-PEG2000), Cas9 RNP complex (pre-formed in nuclease-free duplex buffer), ethanol, 1x PBS (pH 7.4), microfluidic mixer (e.g., NanoAssemblr), dialysis cassettes. Procedure:
- Prepare Lipid Mixture: Dissolve ionizable lipid, DSPC, cholesterol, and PEG-lipid in ethanol at a molar ratio of 50:10:38.5:1.5. Total lipid concentration: 12 mM.
- Prepare Aqueous Phase: Dilute pre-formed Cas9 RNP complex in 50 mM sodium acetate buffer, pH 4.0, to a final concentration of 100 μg/mL.
- Microfluidic Mixing: Use a staggered herringbone mixer. Set the flow rate ratio (aqueous:organic) to 3:1 (total flow rate 12 mL/min). Combine streams in the mixer to form particles.
- Buffer Exchange & Dialysis: Immediately dilute the formed LNP suspension with 1x PBS (pH 7.4) by 2x. Transfer to a dialysis cassette (MWCO 20kDa) and dialyze against 1L of 1x PBS for 4 hours at 4°C, with one buffer change.
- Characterization: Measure particle size and PDI by DLS, zeta potential by electrophoretic light scattering, and RNP encapsulation efficiency using a Ribogreen assay.
Protocol 3.2: Preparation of PEI-based Polyplexes with Cas9 RNP Objective: Form stable, positively charged polyplexes via electrostatic complexation. Materials: Branched Polyethyleneimine (PEI, 25 kDa), Cas9 RNP complex, HEPES-buffered glucose (HBG, pH 7.4), sterile filters (0.22 μm). Procedure:
- Polymer Solution: Prepare a PEI stock solution at 1 mg/mL in HBG, filter sterilize.
- Complexation: Dilute Cas9 RNP to the desired concentration in HBG. Add the appropriate volume of PEI solution to achieve the desired N/P ratio (molar ratio of polymer nitrogen to RNP phosphate). Typical N/P ratios range from 10 to 30.
- Vortex & Incubate: Vortex the mixture immediately for 10 seconds and incubate at room temperature for 30 minutes to allow polyplex formation.
- Characterization: Assess particle size and zeta potential by DLS. Analyze complexation by gel retardation assay (agarose gel electrophoresis).
Protocol 3.3: Conjugation of Cas9 RNP to PEGylated Gold Nanoparticles Objective: Chemically conjugate pre-assembled Cas9 RNP to the surface of AuNPs. Materials: 15 nm PEGylated AuNPs (with carboxylate-terminated PEG), Cas9 RNP, EDC, Sulfo-NHS, MES buffer (pH 6.0), quenching buffer (e.g., 1M ethanolamine, pH 8.5). Procedure:
- Activate AuNP Carboxyl Groups: Mix 1 mL of AuNPs (OD~5) with 10 mM EDC and 25 mM Sulfo-NHS in MES buffer. React for 20 minutes at RT with gentle agitation.
- Purify: Remove excess crosslinkers via centrifugation (12,000g, 15 min) and resuspend in conjugation buffer (e.g., 1x PBS, pH 7.4).
- RNP Conjugation: Incubate the activated AuNPs with Cas9 RNP (molar excess of RNP) for 2 hours at 4°C under gentle rotation.
- Quench Reaction: Add quenching buffer to a final concentration of 50 mM and incubate for 30 minutes to block unreacted sites.
- Purification: Centrifuge to remove unbound RNP (12,000g, 20 min). Resuspend conjugate in storage buffer (PBS with 0.01% Tween-20). Verify conjugation via SDS-PAGE and UV-Vis spectroscopy.
4. Visualization of Key Pathways and Workflows
Diagram 1: Comparative Intracellular Delivery Pathways
Diagram 2: Experimental Workflow for Comparative Analysis
5. The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for RNP-NP Delivery Research
| Item | Function/Application | Example Product/Catalog |
|---|---|---|
| Purified Cas9 Protein | Core nuclease component for RNP assembly. | TruCut HiFi Cas9 Protein (Thermo), Alt-R S.p. Cas9 Nuclease (IDT). |
| Synthetic sgRNA | Guides Cas9 to specific genomic locus. | Alt-R CRISPR-Cas9 sgRNA (IDT), Synthego sgRNA. |
| Ionizable Lipid | Critical LNP component for endosomal escape. | DLin-MC3-DMA (MedKoo), SM-102 (Avanti). |
| Branched Polyethylenimine (PEI) | Gold standard cationic polymer for complexation. | Linear PEI 25kDa (Polysciences), JetPEI (Polyplus). |
| Functionalized Gold Nanoparticles | Ready-to-conjugate AuNP cores. | 15nm AuNP, PEG-Carboxyl (Cytodiagnostics). |
| Microfluidic Mixer | For reproducible, scalable LNP formulation. | NanoAssemblr (Precision NanoSystems). |
| Dynamic Light Scattering (DLS) Instrument | Measures nanoparticle size (hydrodynamic diameter) and polydispersity (PDI). | Zetasizer Nano (Malvern Panalytical). |
| Ribogreen Assay Kit | Quantifies encapsulated/loaded nucleic acid (sgRNA) payload. | Quant-iT RiboGreen (Thermo). |
| T7 Endonuclease I | Detects indel mutations after genome editing. | T7E1 (NEB). |
| Cell Viability Assay Kit | Assesses cytotoxicity of nanoparticle formulations (e.g., MTT, CCK-8). | CellTiter-Glo (Promega). |
Application Notes
The selection of a CRISPR-Cas9 ribonucleoprotein (RNP) delivery method is a critical determinant of experimental and therapeutic success. This note provides a comparative analysis of nanoparticle (NP)-based delivery against electroporation and viral vectors, contextualized within a thesis focused on advancing nanoparticle-RNP platforms for precise genetic engineering.
1. Mechanism & Key Characteristics:
- Nanoparticle (e.g., Lipid Nanoparticle - LNP) RNP Delivery: A synthetic, chemically defined system. Cas9 RNP complexes are encapsulated within or complexed with engineered lipid or polymer nanoparticles. Delivery is primarily through endocytosis, followed by endosomal escape to release the RNP into the cytosol.
- Electroporation/Nucleofection: A physical method applying an external electrical field to transiently permeabilize the cell membrane, allowing direct RNP entry into the cytosol.
- Viral Methods (e.g., AAV, Lentivirus): A biological method using engineered viruses. For RNP delivery, the viral genome typically encodes the Cas9 protein and gRNA, which are expressed intracellularly. Direct viral delivery of pre-formed RNP is less common but under research.
2. Comparative Advantages & Limitations:
Table 1: High-Level Comparison of RNP Delivery Modalities
| Feature | Nanoparticle (LNP) Delivery | Electroporation | Viral Vector (AAV) |
|---|---|---|---|
| Primary Mechanism | Endocytosis & Endosomal Escape | Membrane Permeabilization | Viral Transduction & Expression |
| Delivery Format | Pre-formed RNP | Pre-formed RNP | DNA-Encoded (typically) |
| Typical Transfection Efficiency | 70-95% in vitro (cell-dependent) | 80-99% in vitro | 30-90% (serotype-dependent) |
| Cellular Toxicity | Low to Moderate (depends on formulation) | High (affects cell viability) | Low (but immunogenic) |
| In Vivo Applicability | Excellent (systemic or local) | Limited (ex vivo mainly) | Good (but pre-existing immunity) |
| Immunogenicity | Low (modifiable surface) | None (physical method) | High (neutralizing antibodies) |
| Loading Capacity | High (can carry large RNPs + donors) | High (direct cytosolic entry) | Low (~4.7 kb for AAV) |
| Manufacturing | Scalable, synthetic | Instrument-based | Complex biological production |
| Cost per Experiment | Moderate | Low (capital investment high) | High |
| Therapeutic Timeline | Rapid action (hours) | Rapid action (hours) | Slow (requires transcription/translation) |
| Risk of Off-Target Integration | Lowest (RNP degrades quickly) | Low | High (viral genome integration risk) |
Table 2: Quantitative Performance Metrics in Primary T-Cell Editing (Representative Data)
| Metric | Gold Nanoparticle RNP | Electroporation (Neon) | LNP RNP |
|---|---|---|---|
| Viability (Day 3 post-edit) | 85% ± 5% | 65% ± 10% | 90% ± 4% |
| Knockout Efficiency (% CD3+) | 60% ± 8% | 85% ± 5% | 78% ± 6% |
| Proliferation Capacity | Maintained | Reduced | Enhanced |
| Cytokine Release (IL-6) | Baseline | Elevated | Baseline |
| Process Time | 2 hours | <1 hour | 3 hours (incubation) |
Experimental Protocols
Protocol 1: CRISPR-Cas9 RNP Knockout in Primary Human T-cells Using Lipid Nanoparticles Objective: To achieve high-efficiency gene knockout in primary T-cells with high viability using LNPs.
- RNP Complex Formation: Incubate 6 µg of purified Cas9 protein with 3 µg of chemically synthesized sgRNA (targeting gene of interest, e.g., PD-1) in a duplex buffer for 10 min at 25°C to form RNP complexes.
- LNP Formulation: Use a microfluidic device to mix an ethanolic lipid phase (ionizable lipid: DOPE: Cholesterol: PEG-lipid = 50:10:38.5:1.5 mol%) with an aqueous phase containing the formed RNP complexes in citrate buffer (pH 4.0). Dialyze the resulting LNP suspension against 1X PBS (pH 7.4) for 2 hours.
- T-cell Culture: Isolate and activate primary human T-cells using CD3/CD28 beads for 48 hours in IL-2 supplemented media.
- Transfection: Wash activated T-cells. Add LNP-RNP at a final lipid concentration of 200 nM to 1e6 cells/mL in a 24-well plate. Incubate for 6 hours at 37°C.
- Recovery & Analysis: Replace LNP-containing media with fresh complete media. Analyze editing efficiency at 72-96 hours post-transfection via flow cytometry (for surface proteins) or next-generation sequencing (NGS) of the target locus.
Protocol 2: Electroporation of CRISPR-Cas9 RNP into Hematopoietic Stem/Progenitor Cells (HSPCs) Objective: To efficiently edit hard-to-transfect HSPCs using a clinically relevant electroporation system.
- RNP Complex Formation: Assemble Cas9 protein and sgRNA at a 1:2 molar ratio (e.g., 6 nmol Cas9: 12 nmol sgRNA) in PBS for 10 min at room temperature.
- Cell Preparation: Purify CD34+ HSPCs. Wash cells twice in PBS and resuspend in the recommended electroporation buffer (e.g., P3 buffer) at 1e7 cells/mL.
- Electroporation: Mix 10 µL of cell suspension (1e5 cells) with 2 µL of pre-assembled RNP (final ~10 µM) in a 16-well electroporation strip. Electroporate using a 4D-Nucleofector (Program DZ-100). Immediately add 80 µL of pre-warmed culture medium.
- Recovery: Transfer cells to a collagen-coated plate with complete stem cell media. Analyze editing and colony-forming unit (CFU) assays after 7-14 days.
Visualizations
Diagram 1: Endosomal Escape Pathway for Nanoparticle-Delivered RNP
Diagram 2: Workflow Comparison for Three RNP Delivery Methods
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for Nanoparticle-Based RNP Delivery Research
| Reagent/Material | Function & Rationale |
|---|---|
| Purified Cas9 Protein (e.g., SpyCas9) | The effector enzyme. High-purity, endotoxin-free protein is critical for RNP assembly and to minimize immune activation. |
| Chemically Modified sgRNA (e.g., 2'-O-methyl, phosphorothioate) | Enhances stability against nucleases, increases RNP half-life, and can improve editing efficiency. |
| Ionizable Cationic Lipid (e.g., DLin-MC3-DMA, ALC-0315) | Core component of LNPs. Protonates in acidic endosomes, enabling membrane disruption and RNP escape. |
| Helper Lipid (e.g., DOPE, DSPC) | Supports the formation and stability of the lipid bilayer and can promote endosomal escape via fusogenic properties. |
| PEGylated Lipid (e.g., DMG-PEG2000) | Shields the LNP surface, improves colloidal stability, reduces non-specific binding, and modulates pharmacokinetics in vivo. |
| Microfluidic Mixer (e.g., NanoAssemblr, Staggered Herringbone) | Enables reproducible, scalable, and rapid mixing for the formation of homogeneous, small-diameter LNPs. |
| Dynamic Light Scattering (DLS) Instrument | Measures critical quality attributes of LNPs: hydrodynamic diameter (size), polydispersity index (PDI), and zeta potential. |
| Endotoxin-Free Buffers & Consumables | Essential for in vivo applications and sensitive primary cell work to prevent confounding inflammatory responses. |
Within the broader thesis on CRISPR-Cas9 ribonucleoprotein (RNP) delivery via engineered nanoparticles (NPs), profiling immunogenicity and safety is paramount. The goal is to achieve efficient gene editing while minimizing undesirable immune activation that could lead to acute inflammatory responses, reduced editing efficacy, or long-term adverse effects. This document outlines standardized protocols for assessing innate immune activation by nanoparticle-delivered RNPs and for evaluating the establishment of long-term immune tolerance to both the delivery vehicle and the therapeutic cargo.
Key Focus Areas:
- Innate Immune Sensing: NPs and residual nucleic acids (e.g., guide RNA fragments, bacterial-derived Cas9 mRNA) can be detected by pattern recognition receptors (PRRs) like Toll-like receptors (TLRs) and cytosolic sensors (e.g., RIG-I, cGAS), leading to type I interferon (IFN) and pro-inflammatory cytokine production.
- Safety & Tolerance: Chronic immune activation is detrimental. The protocols assess acute cytokine release, leukocyte infiltration, and the induction of regulatory mechanisms that may promote antigen-specific tolerance to Cas9 and the NP components.
Key Experimental Protocols
Protocol 2.1: In Vitro Profiling of Innate Immune Activation in Primary Human Immune Cells
Objective: To quantitatively measure cytokine/chemokine release and cell surface activation markers following treatment with CRISPR-Cas9 RNP NPs.
Materials:
- Primary human peripheral blood mononuclear cells (PBMCs) or purified plasmacytoid dendritic cells (pDCs) and monocytes.
- Test articles: LNP- or polymer-based NPs loaded with Cas9 RNP, control NPs (empty, scrambled gRNA), positive controls (LPS, CpG ODN).
- Cell culture media (e.g., RPMI-1640 + 10% FBS).
- Multi-analyte cytokine detection kit (e.g., Luminex or MSD) targeting IFN-α, IFN-β, IL-6, TNF-α, IL-1β, IP-10.
- Flow cytometry antibodies: CD14, CD16, CD86, HLA-DR, CD80, CD11c, BDCA-2.
Methodology:
- Isolate PBMCs from healthy donor buffy coats using density gradient centrifugation.
- Seed cells in 96-well plates at 2x10^5 cells/well.
- Treat cells with a dose range of RNP-NPs (e.g., 0.1, 1, 10 µg/mL total lipid/polymer), controls, and media alone. Use n=4-6 replicates.
- Incubate for 6h (early activation markers), 24h, and 48h (cytokine secretion).
- Supernatant Collection: Harvest supernatant, centrifuge to remove debris, and store at -80°C. Analyze cytokines using a multiplex immunoassay per manufacturer's protocol.
- Cell Analysis: Harvest cells, stain with viability dye and antibody panels for flow cytometry. Analyze activation marker (e.g., CD86) MFI increases on target populations (monocytes, pDCs).
Data Analysis: Normalize data to media control. Calculate fold-change and statistical significance (e.g., one-way ANOVA).
Protocol 2.2: In Vivo Assessment of Acute Inflammatory Response and Editing Efficiency
Objective: To correlate systemic cytokine responses and local immune cell infiltration with target tissue editing efficiency in a murine model.
Materials:
- C57BL/6 mice (or relevant disease model).
- RNP-NPs targeting a genomic safe harbor locus (e.g., Rosa26).
- ELISA kits for murine IFN-β, IL-6, KC/GRO.
- Tissue dissociation kits, flow cytometry antibodies for murine CD45, Ly6C, Ly6G, F4/80, CD11b.
- Next-generation sequencing (NGS) reagents for Indel analysis.
Methodology:
- Administer RNP-NPs systemically (e.g., IV injection) at the therapeutic dose. Include PBS and empty NP controls (n=5-8/group).
- Serum Collection: Retro-orbitally bleed mice at 2h, 6h, and 24h post-injection. Isolate serum, analyze cytokines via ELISA.
- Tissue Harvest: At 48h (acute inflammation) and 2 weeks (editing efficiency), euthanize mice. Collect target tissue (e.g., liver), spleen, and draining lymph nodes.
- Immune Profiling: Process tissues into single-cell suspensions. Perform flow cytometry to quantify innate immune cell (neutrophil, monocyte, macrophage) infiltration in target tissue.
- Editing Analysis: Isolate genomic DNA from target tissue. Amplify the target locus by PCR and perform NGS to determine indel frequency.
Protocol 2.3: Evaluating Long-Term Tolerance to Cas9 and NP Components
Objective: To assess antigen-specific T-cell tolerance and humoral responses after repeated administration.
Materials:
- Mice from Protocol 2.2.
- Recombinant Cas9 protein, NP component (e.g., ionizable lipid).
- ELISpot kits for IFN-γ and IL-10.
- ELISA for anti-Cas9 and anti-PEG antibodies.
Methodology:
- Re-challenge: At 4 weeks post-initial dose, administer a sub-therapeutic booster dose of the same RNP-NPs or the antigenic components alone.
- Humoral Response: Measure serum levels of anti-Cas9 and anti-NP component IgG/IgM antibodies via ELISA at baseline, pre-boost, and 7 days post-boost.
- Cellular Response: 10 days post-boost, isolate splenocytes. Perform Cas9-specific T-cell recall assays using ELISpot. Stimulate splenocytes with Cas9 peptide pools; quantify IFN-γ (effector) and IL-10 (regulatory) spot-forming units.
- Histopathology: Perform H&E staining on target organs (liver, spleen) to assess chronic inflammation or granuloma formation.
Data Presentation
Table 1: In Vitro Cytokine Profile of Primary Human PBMCs Treated with RNP-NPs (24h)
| Treatment (10 µg/mL) | IFN-α (pg/mL) | IFN-β (pg/mL) | IL-6 (pg/mL) | TNF-α (pg/mL) |
|---|---|---|---|---|
| Media Control | 5 ± 2 | 10 ± 3 | 15 ± 5 | 8 ± 2 |
| Empty NPs | 20 ± 8 | 45 ± 12 | 120 ± 30 | 55 ± 15 |
| RNP-NPs (Polymer A) | 250 ± 45 | 520 ± 80 | 1500 ± 200 | 800 ± 110 |
| RNP-NPs (LNP B) | 50 ± 15 | 180 ± 40 | 600 ± 90 | 300 ± 50 |
| Positive Control (CpG) | 1250 ± 300 | 2800 ± 500 | 3500 ± 600 | 1900 ± 400 |
Table 2: In Vivo Correlation of Inflammation and Editing (Murine Liver, 48h & 2wk)
| Treatment Group | Serum IL-6 @6h (pg/mL) | Liver Infiltrating Neutrophils (% of CD45+) | Indel Frequency @2wk (%) |
|---|---|---|---|
| PBS | 10 ± 3 | 2.1 ± 0.5 | N/A |
| Empty LNP | 85 ± 20 | 8.5 ± 1.8 | N/A |
| RNP-LNP (Standard) | 450 ± 75 | 25.4 ± 4.2 | 45 ± 6 |
| RNP-LNP (Engineered)* | 120 ± 30 | 9.8 ± 2.1 | 52 ± 7 |
*Engineered for reduced immunogenicity.
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Profiling |
|---|---|
| Recombinant Cas9 Protein | Positive control for immune assays; antigen for tolerance studies and antibody detection. |
| TLR Inhibitors (e.g., TLR7/8 inhibitor CU-CPT9a) | To mechanistically dissect innate sensing pathways in vitro. |
| PEGylated Lipids | Common NP component; monitor anti-PEG antibodies as a marker of humoral immunogenicity. |
| Cyclic GMP-AMP (cGAMP) | Positive control for activating the cGAS-STING cytosolic DNA sensing pathway. |
| High-Sensitivity Multiplex Cytokine Assays (MSD/Luminex) | Quantify broad panels of pro- and anti-inflammatory cytokines from small sample volumes. |
| Cas9 Peptide MegaPools (Overlapping 15-mers) | To comprehensively map Cas9-specific T-cell responses in ELISpot/T-cell assays. |
| Endotoxin-Free Labware & Reagents | Critical to ensure measured immune responses are due to NPs, not contaminating LPS. |
Visualizations
Diagram 1: Innate Immune Sensing Pathways for RNP-NPs
Diagram 2: Immunogenicity & Tolerance Profiling Workflow
Conclusion
Nanoparticle-mediated delivery of CRISPR-Cas9 RNP complexes represents a rapidly maturing field that addresses critical limitations of traditional genome editing delivery methods. By combining the inherent safety and precision of RNPs with the protective, targeted, and tunable nature of nanocarriers, this synergy offers a powerful pathway toward clinically viable therapeutics. Key takeaways include the superiority of LNPs for systemic delivery, the importance of endosomal escape mechanisms, and the necessity of rigorous off-target and immunogenicity profiling. Future directions point towards the development of smart, stimuli-responsive nanoparticles, advanced in vivo targeting strategies, and the convergence with mRNA delivery technologies. As formulation science and biological understanding advance, nanoparticle-RNP platforms are poised to accelerate the translation of CRISPR-based therapies from preclinical research to transformative clinical applications in genetic disorders, oncology, and beyond.